MicrobiotaProcess Group2 vs Group6 (v1)
gene_x 0 like s 1024 view s
Tags: plot, packages, R, processing, pipeline
https://bioconductor.org/packages/release/bioc/vignettes/MicrobiotaProcess/inst/doc//MicrobiotaProcess.html
-
code of MicrobiotaProcess_Group2_vs_Group6.R
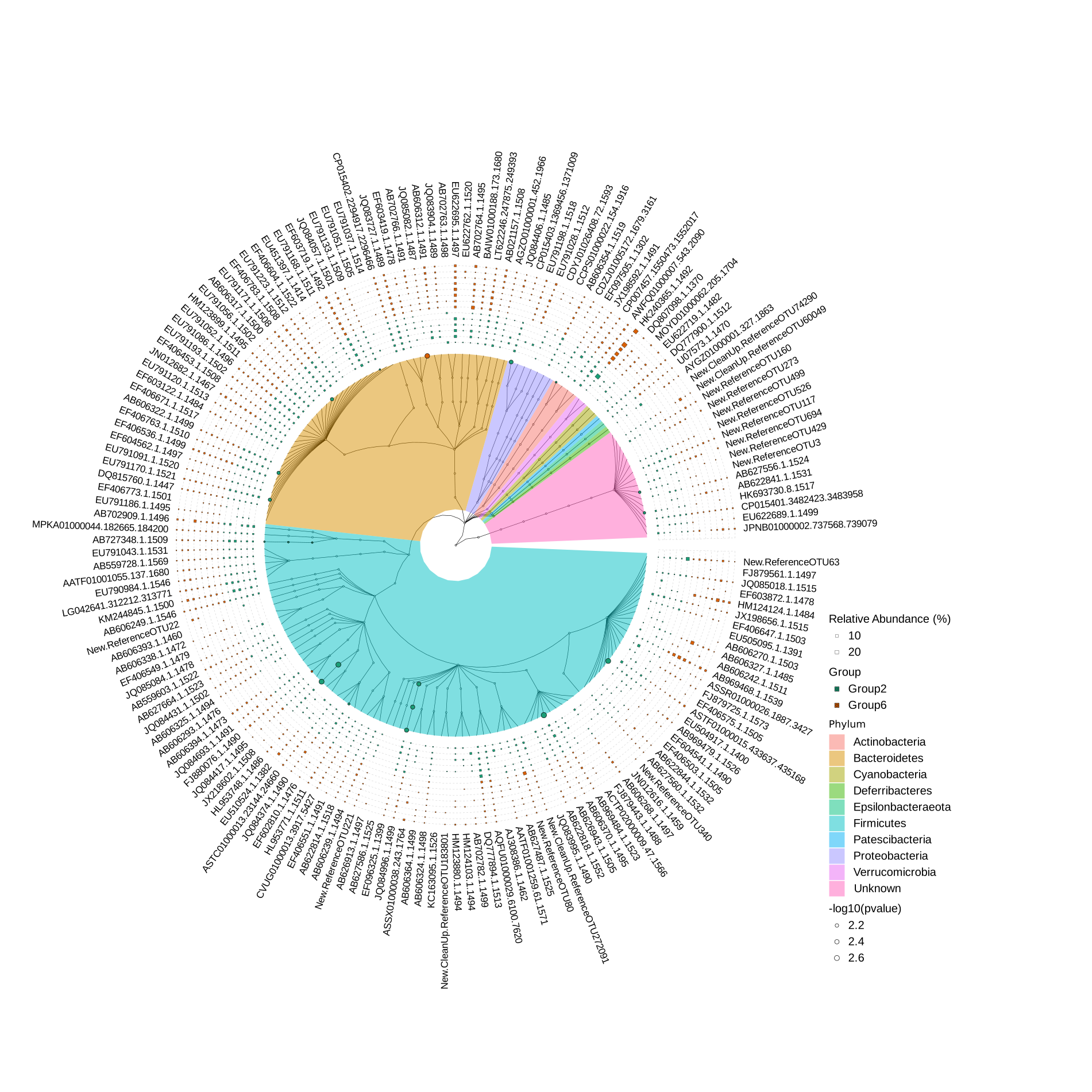
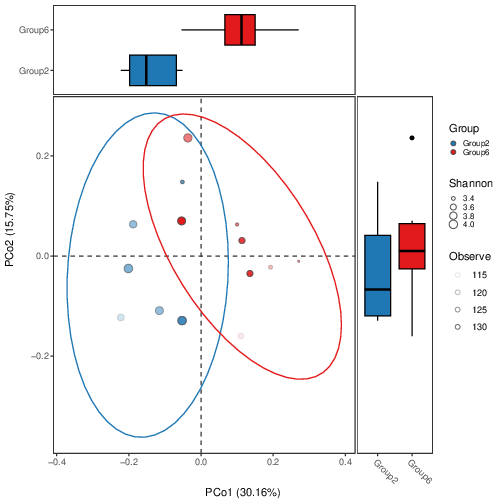
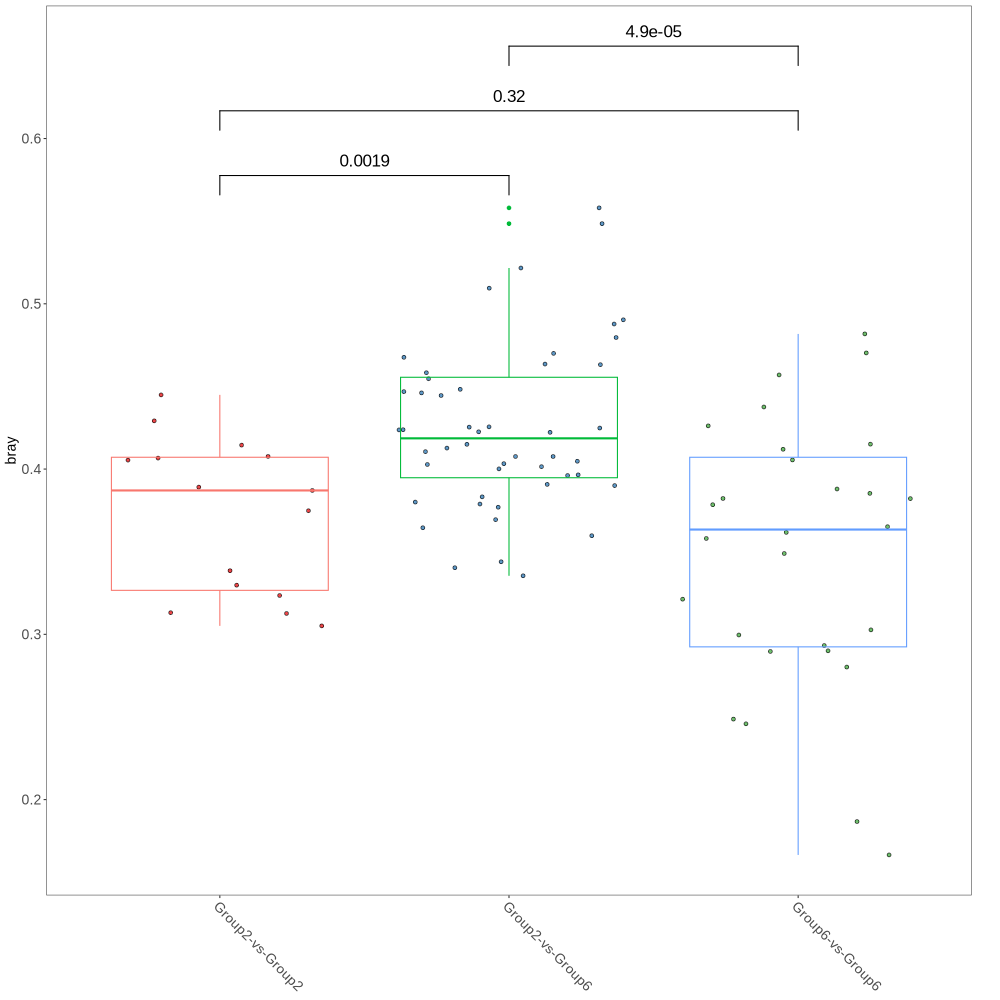
# https://bioconductor.org/packages/release/bioc/vignettes/MicrobiotaProcess/inst/doc//MicrobiotaProcess.html# -----------------------------------# ---- prepare the R environment ----#Rscript MicrobiotaProcess.R#NOTE: exit R script, then login again R-environment; rm -rf Phyloseq*_cachermarkdown::render('Phyloseq.Rmd',output_file='Phyloseq.html')# -----------------------------# ---- 3.1. bridges other tools##https://github.com/YuLab-SMU/MicrobiotaProcess##https://www.bioconductor.org/packages/release/bioc/vignettes/MicrobiotaProcess/inst/doc/MicrobiotaProcess.html##https://chiliubio.github.io/microeco_tutorial/intro.html#framework##https://yiluheihei.github.io/microbiomeMarker/reference/plot_cladogram.html#BiocManager::install("MicrobiotaProcess")#install.packages("microeco")#install.packages("ggalluvial")#install.packages("ggh4x")library(MicrobiotaProcess)library(microeco)library(ggalluvial)library(ggh4x)library(gghalves)## Convert the phyloseq object to a MicrobiotaProcess object#mp <- as.MicrobiotaProcess(ps.ng.tax)#mt <- phyloseq2microeco(ps.ng.tax) #--> ERROR#abundance_table <- mt$abun_table#taxonomy_table <- mt$tax_table#ps.ng.tax_abund <- phyloseq::filter_taxa(ps.ng.tax, function(x) sum(x > total*0.01) > 0, TRUE)#ps.ng.tax_most = phyloseq::filter_taxa(ps.ng.tax_rel, function(x) mean(x) > 0.001, TRUE)##OPTION1 (NOT_USED): take all samples, prepare ps.ng.tax_abund --> mpse_abund##mpse <- ps.ng.tax %>% as.MPSE()#mpse_abund <- ps.ng.tax_abund %>% as.MPSE()##OPTION2 (USED!): take partial samples, prepare ps.ng.tax or ps.ng.tax_abund (2 replacements!)--> ps.ng.tax_sel --> mpse_abundps.ng.tax_sel <- ps.ng.tax_abund##otu_table(ps.ng.tax_sel) <- otu_table(ps.ng.tax)[,c("1","2","5","6","7", "15","16","17","18","19","20", "29","30","31","32", "40","41","42","43","44","46")]##NOTE: Only choose Group2, Group4, Group6, Group8#> ps.ng.tax_sel#otu_table() OTU Table: [ 37465 taxa and 29 samples ]#sample_data() Sample Data: [ 29 samples by 10 sample variables ]#tax_table() Taxonomy Table: [ 37465 taxa by 7 taxonomic ranks ]#phy_tree() Phylogenetic Tree: [ 37465 tips and 37461 internal nodes ]#-Group4: "21","22","23","24","25","26","27","28",#-Group8: , "47","48","49","50","52","53","55"otu_table(ps.ng.tax_sel) <- otu_table(ps.ng.tax_abund)[,c("8","9","10","12","13","14", "33","34","35","36","37","38","39","51")]mpse_abund <- ps.ng.tax_sel %>% as.MPSE()# A MPSE-tibble (MPSE object) abstraction: 2,352 × 20# NOTE mpse_abund contains 20 variables: OTU, Sample, Abundance, BarcodeSequence, LinkerPrimerSequence, FileInput, Group,# Sex_age <chr>, pre_post_stroke <chr>, Conc <dbl>, Vol_50ng <dbl>, Vol_PCR <dbl>, Description <chr>,# Domain <chr>, Phylum <chr>, Class <chr>, Order <chr>, Family <chr>, Genus <chr>, Species <chr># -----------------------------------# ---- 3.2. alpha diversity analysis# Rarefied species richness + RareAbundancempse_abund %<>% mp_rrarefy()# 'chunks' represent the split number of each sample to calculate alpha# diversity, default is 400. e.g. If a sample has total 40000# reads, if chunks is 400, it will be split to 100 sub-samples# (100, 200, 300,..., 40000), then alpha diversity index was# calculated based on the sub-samples.# '.abundance' the column name of abundance, if the '.abundance' is not be# rarefied calculate rarecurve, user can specific 'force=TRUE'.mpse_abund %<>%mp_cal_rarecurve(.abundance = RareAbundance,chunks = 400)# The RareAbundanceRarecurve column will be added the colData slot# automatically (default action="add")#NOTE mpse_abund contains 22 varibles = 20 varibles + RareAbundance <dbl> + RareAbundanceRarecurve <list># default will display the confidence interval around smooth.# se=TRUE# NOTE that two colors #c("#00A087FF", "#3C5488FF") for .group = pre_post_stroke; four colors c("#1f78b4", "#33a02c", "#e31a1c", "#6a3d9a") for .group = Group;p1 <- mpse_abund %>%mp_plot_rarecurve(.rare = RareAbundanceRarecurve,.alpha = Observe,)p2 <- mpse_abund %>%mp_plot_rarecurve(.rare = RareAbundanceRarecurve,.alpha = Observe,.group = Group) +scale_color_manual(values=c("#1f78b4", "#e31a1c")) +scale_fill_manual(values=c("#1f78b4", "#e31a1c"), guide="none")# combine the samples belong to the same groups if plot.group=TRUEp3 <- mpse_abund %>%mp_plot_rarecurve(.rare = RareAbundanceRarecurve,.alpha = "Observe",.group = Group,plot.group = TRUE) +scale_color_manual(values=c("#1f78b4", "#e31a1c")) +scale_fill_manual(values=c("#1f78b4", "#e31a1c"),guide="none")png("rarefaction_of_samples_or_groups.png", width=1080, height=600)p1 + p2 + p3dev.off()# ------------------------------------------# 3.3. calculate alpha index and visualizationlibrary(ggplot2)library(MicrobiotaProcess)mpse_abund %<>%mp_cal_alpha(.abundance=RareAbundance)mpse_abund#NOTE mpse_abund contains 28 varibles = 22 varibles + Observe <dbl>, Chao1 <dbl>, ACE <dbl>, Shannon <dbl>, Simpson <dbl>, Pielou <dbl>f1 <- mpse_abund %>%mp_plot_alpha(.group=Group,.alpha=c(Observe, Chao1, ACE, Shannon, Simpson, Pielou)) +scale_fill_manual(values=c("#1f78b4", "#e31a1c"), guide="none") +scale_color_manual(values=c("#1f78b4", "#e31a1c"), guide="none")f2 <- mpse_abund %>%mp_plot_alpha(.alpha=c(Observe, Chao1, ACE, Shannon, Simpson, Pielou))#ps.ng.tax_sel contais only pre samples --> f1 cannot be generated!png("alpha_diversity_comparison.png", width=1400, height=600)f1 / f2dev.off()# -------------------------------------------# 3.4. The visualization of taxonomy abundance (Class)mpse_abund %<>%mp_cal_abundance( # for each samples.abundance = RareAbundance) %>%mp_cal_abundance( # for each groups.abundance=RareAbundance,.group=Group)mpse_abund#NOTE mpse_abund contains 29 varibles = 28 varibles + RelRareAbundanceBySample <dbl># visualize the relative abundance of top 20 phyla for each sample.# .group=time,p1 <- mpse_abund %>%mp_plot_abundance(.abundance=RareAbundance,taxa.class = Class,topn = 20,relative = TRUE)# visualize the abundance (rarefied) of top 20 phyla for each sample.# .group=time,p2 <- mpse_abund %>%mp_plot_abundance(.abundance=RareAbundance,taxa.class = Class,topn = 20,relative = FALSE)png("relative_abundance_and_abundance.png", width= 1200, height=600) #NOT PRODUCED!p1 / p2dev.off()#----h1 <- mpse_abund %>%mp_plot_abundance(.abundance = RareAbundance,.group = Group,taxa.class = Class,relative = TRUE,topn = 20,geom = 'heatmap',features.dist = 'euclidean',features.hclust = 'average',sample.dist = 'bray',sample.hclust = 'average')h2 <- mpse_abund %>%mp_plot_abundance(.abundance = RareAbundance,.group = Group,taxa.class = Class,relative = FALSE,topn = 20,geom = 'heatmap',features.dist = 'euclidean',features.hclust = 'average',sample.dist = 'bray',sample.hclust = 'average')# the character (scale or theme) of figure can be adjusted by set_scale_theme# refer to the mp_plot_distpng("relative_abundance_and_abundance_heatmap.png", width= 1200, height=600)aplot::plot_list(gglist=list(h1, h2), tag_levels="A")dev.off()# visualize the relative abundance of top 20 class for each .group (Group)p3 <- mpse_abund %>%mp_plot_abundance(.abundance=RareAbundance,.group=Group,taxa.class = Class,topn = 20,plot.group = TRUE)# visualize the abundance of top 20 phyla for each .group (time)p4 <- mpse_abund %>%mp_plot_abundance(.abundance=RareAbundance,.group= Group,taxa.class = Class,topn = 20,relative = FALSE,plot.group = TRUE)png("relative_abundance_and_abundance_groups.png", width= 1000, height=1000)p3 / p4dev.off()# ---------------------------# 3.5. Beta diversity analysis# ---------------------------------------------# 3.5.1 The distance between samples or groups# standardization# mp_decostand wraps the decostand of vegan, which provides# many standardization methods for community ecology.# default is hellinger, then the abundance processed will# be stored to the assays slot.mpse_abund %<>%mp_decostand(.abundance=Abundance)mpse_abund#NOTE mpse_abund contains 30 varibles = 29 varibles + hellinger <dbl># calculate the distance between the samples.# the distance will be generated a nested tibble and added to the# colData slot.mpse_abund %<>% mp_cal_dist(.abundance=hellinger, distmethod="bray")mpse_abund#NOTE mpse_abund contains 31 varibles = 30 varibles + bray <list># mp_plot_dist provides there methods to visualize the distance between the samples or groups# when .group is not provided, the dot heatmap plot will be returnp1 <- mpse_abund %>% mp_plot_dist(.distmethod = bray)png("distance_between_samples.png", width= 1000, height=1000)p1dev.off()# when .group is provided, the dot heatmap plot with group information will be return.p2 <- mpse_abund %>% mp_plot_dist(.distmethod = bray, .group = Group)# The scale or theme of dot heatmap plot can be adjusted using set_scale_theme function.p2 %>% set_scale_theme(x = scale_fill_manual(values=c("#1f78b4", "#e31a1c"), #c("orange", "deepskyblue"),guide = guide_legend(keywidth = 1,keyheight = 0.5,title.theme = element_text(size=8),label.theme = element_text(size=6))),aes_var = Group # specific the name of variable) %>%set_scale_theme(x = scale_color_gradient(guide = guide_legend(keywidth = 0.5, keyheight = 0.5)),aes_var = bray) %>%set_scale_theme(x = scale_size_continuous(range = c(0.1, 3),guide = guide_legend(keywidth = 0.5, keyheight = 0.5)),aes_var = bray)png("distance_between_samples_with_group_info.png", width= 1000, height=1000)p2dev.off()# when .group is provided and group.test is TRUE, the comparison of different groups will be returned# Assuming p3 is a ggplot object after mp_plot_dist callp3 <- mpse_abund %>%mp_plot_dist(.distmethod = bray, .group = Group, group.test = TRUE, textsize = 6) +theme(axis.title.x = element_text(size = 14), # Customize x-axis label face = "bold"axis.title.y = element_text(size = 14), # Customize y-axis labelaxis.text.x = element_text(size = 14), # Customize x-axis ticksaxis.text.y = element_text(size = 14) # Customize y-axis ticks)# Save the plot with the new theme settingspng("Comparison_of_Bray_Distances.png", width = 1000, height = 1000)print(p3) # Ensure that p3 is explicitly printed in the devicedev.off()# Extract Bray-Curtis Distance Values and save them in a Excel-table.library(dplyr)library(tidyr)library(openxlsx)# Define the sample numbers vectorsample_numbers <- c("8", "9", "10", "12", "13", "14", "33", "34", "35", "36", "37", "38", "39", "51")# Consolidate the list of tibbles using the actual sample numbersbray_data <- bind_rows(lapply(seq_along(mpse_abund$bray), function(i) {tibble(Sample1 = sample_numbers[i], # Use actual sample numberSample2 = mpse_abund$bray[[i]]$braySampley,BrayDistance = mpse_abund$bray[[i]]$bray)}),.id = "PairID")# Print the data frame to check the outputprint(bray_data)# Write the data frame to an Excel filewrite.xlsx(bray_data, file = "Bray_Curtis_Distances.xlsx")#DELETE the column "PairID" in Excel file# -----------------------# 3.5.2 The PCoA analysis#install.packages("corrr")library(corrr)#install.packages("ggside")library(ggside)mpse_abund %<>%mp_cal_pcoa(.abundance=hellinger, distmethod="bray")# The dimensions of ordination analysis will be added the colData slot (default).mpse_abundmpse_abund %>% print(width=380, n=2)#NOTE mpse_abund contains 34 varibles = 31 varibles + `PCo1 (30.16%)` <dbl>, `PCo2 (15.75%)` <dbl>, `PCo3 (10.53%)` <dbl>#BUG why 36 variables in mpse_abund %>% print(width=380, n=1) [RareAbundanceBySample <list>, RareAbundanceByGroup <list>]#> methods(class=class(mpse_abund))# [1] [ [[<- [<-# [4] $ $<- arrange# [7] as_tibble as.data.frame as.phyloseq#[10] coerce coerce<- colData<-#[13] distinct filter group_by#[16] left_join mp_adonis mp_aggregate_clade#[19] mp_aggregate mp_anosim mp_balance_clade#[22] mp_cal_abundance mp_cal_alpha mp_cal_cca#[25] mp_cal_clust mp_cal_dca mp_cal_dist#[28] mp_cal_nmds mp_cal_pca mp_cal_pcoa#[31] mp_cal_pd_metric mp_cal_rarecurve mp_cal_rda#[34] mp_cal_upset mp_cal_venn mp_decostand#[37] mp_diff_analysis mp_diff_clade mp_envfit#[40] mp_extract_abundance mp_extract_assays mp_extract_dist#[43] mp_extract_feature mp_extract_internal_attr mp_extract_rarecurve#[46] mp_extract_refseq mp_extract_sample mp_extract_taxonomy#[49] mp_extract_tree mp_filter_taxa mp_mantel#[52] mp_mrpp mp_plot_abundance mp_plot_alpha#[55] mp_plot_diff_boxplot mp_plot_diff_res mp_plot_dist#[58] mp_plot_ord mp_plot_rarecurve mp_plot_upset#[61] mp_plot_venn mp_rrarefy mp_select_as_tip#[64] mp_stat_taxa mutate otutree#[67] otutree<- print pull#[70] refsequence refsequence<- rename#[73] rownames<- select show# [ reached getOption("max.print") -- omitted 6 entries ]#see '?methods' for accessing help and source code# We also can perform adonis or anosim to check whether it is significant to the dissimilarities of groups.mpse_abund %<>%mp_adonis(.abundance=hellinger, .formula=~Group, distmethod="bray", permutations=9999, action="add")mpse_abund %>% mp_extract_internal_attr(name=adonis)#NOTE mpse_abund contains 34 varibles, no new variable, it has been saved in mpse_abund and can be extracted with "mpse_abund %>% mp_extract_internal_attr(name='adonis')"#The result of adonis has been saved to the internal attribute !#It can be extracted using this-object %>% mp_extract_internal_attr(name='adonis')#The object contained internal attribute: PCoA ADONIS#Permutation test for adonis under reduced model#Terms added sequentially (first to last)#Permutation: free#Number of permutations: 9999##vegan::adonis2(formula = .formula, data = sampleda, permutations = permutations, method = distmethod)# Df SumOfSqs R2 F Pr(>F)#Group 1 0.23448 0.22659 3.5158 5e-04 ***#Residual 12 0.80032 0.77341#Total 13 1.03480 1.00000#---#Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1# ("1","2","5","6","7", "15","16","17","18","19","20", "29","30","31","32", "40","41","42","43","44","46")#div.df2[div.df2 == "Group1"] <- "aged.post"#div.df2[div.df2 == "Group3"] <- "young.post"#div.df2[div.df2 == "Group5"] <- "aged.post"#div.df2[div.df2 == "Group7"] <- "young.post"# ("8","9","10","12","13","14", "21","22","23","24","25","26","27","28", "33","34","35","36","37","38","39","51", "47","48","49","50","52","53","55")#div.df2[div.df2 == "Group2"] <- "aged.pre"#div.df2[div.df2 == "Group4"] <- "young.pre"#div.df2[div.df2 == "Group6"] <- "aged.pre"#div.df2[div.df2 == "Group8"] <- "young.pre"#Group1: f.aged and post#Group2: f.aged and pre#Group3: f.young and post#Group4: f.young and pre#Group5: m.aged and post#Group6: m.aged and pre#Group7: m.young and post#Group8: m.young and pre#[,c("1","2","5","6","7", "8","9","10","12","13","14")]#[,c("15","16","17","18","19","20", "21","22","23","24","25","26","27","28")]#[,c("29","30","31","32", "33","34","35","36","37","38","39","51")]#[,c("40","41","42","43","44","46", "47","48","49","50","52","53","55")]#For the first set:#a6cee3: This is a light blue color, somewhat pastel and soft.#b2df8a: A soft, pale green, similar to a light lime.#fb9a99: A soft pink, slightly peachy or salmon-like.#cab2d6: A pale purple, reminiscent of lavender or a light mauve.#For the second set:#1f78b4: This is a strong, vivid blue, close to cobalt or a medium-dark blue.#33a02c: A medium forest green, vibrant and leafy.#e31a1c: A bright red, very vivid, similar to fire engine red.#6a3d9a: This would be described as a deep purple, akin to a dark lavender or plum.p1 <- mpse_abund %>%mp_plot_ord(.ord = pcoa,.group = Group,.color = Group,.size = 2.4,.alpha = 1,ellipse = TRUE,show.legend = FALSE # don't display the legend of stat_ellipse) +scale_fill_manual(#values = c("#a6cee3", "#1f78b4", "#b2df8a", "#33a02c", "#fb9a99", "#e31a1c", "#cab2d6", "#6a3d9a"),#values = c("#a6cee3", "#b2df8a", "#fb9a99", "#cab2d6"),values = c("#1f78b4", "#e31a1c"),guide = guide_legend(keywidth=1.6, keyheight=1.6, label.theme=element_text(size=12))) +scale_color_manual(#values=c("#a6cee3", "#1f78b4", "#b2df8a", "#33a02c", "#fb9a99", "#e31a1c", "#cab2d6", "#6a3d9a"),#values = c("#a6cee3", "#b2df8a", "#fb9a99", "#cab2d6"),values = c("#1f78b4", "#e31a1c"),guide = guide_legend(keywidth=1.6, keyheight=1.6, label.theme=element_text(size=12)))#scale_fill_manual(values=c("#FF0000", "#000000", "#0000FF", "#C0C0C0", "#00FF00", "#FFFF00", "#00FFFF", "#FFA500")) +#scale_color_manual(values=c("#FF0000", "#000000", "#0000FF", "#C0C0C0", "#00FF00", "#FFFF00", "#00FFFF", "#FFA500"))#scale_fill_manual(values=c("#00A087FF", "#3C5488FF")) +#scale_color_manual(values=c("#00A087FF", "#3C5488FF"))#png("PCoA.png", width= 1000, height=1000)#svg("PCoA.svg", width= 11, height=10)#svg("PCoA_.svg", width=10, height=10)#svg("PCoA.svg")pdf("PCoA_Group2_vs_Group6.pdf")p1dev.off()#FF0000: Red#000000: Black#0000FF: Blue#C0C0C0: Silver#00FF00: Lime (often referred to simply as Green in web colors)#FFFF00: Yellow#00FFFF: Aqua (also known as Cyan)#FFA500: Orange# The size of point also can be mapped to other variables such as Observe, or Shannon# Then the alpha diversity and beta diversity will be displayed simultaneously.p2 <- mpse_abund %>%mp_plot_ord(.ord = pcoa,.group = Group,.color = Group,.size = Shannon,.alpha = Observe,ellipse = TRUE,show.legend = FALSE # don't display the legend of stat_ellipse) +scale_fill_manual(values = c("#1f78b4", "#e31a1c"), #only needs four colors.#values = c("#FF0000", "#000000", "#0000FF", "#C0C0C0", "#00FF00", "#FFFF00", "#00FFFF", "#FFA500"),guide = guide_legend(keywidth=0.6, keyheight=0.6, label.theme=element_text(size=8))) +scale_color_manual(values = c("#1f78b4", "#e31a1c"), #only needs four colors.#values=c("#FF0000", "#000000", "#0000FF", "#C0C0C0", "#00FF00", "#FFFF00", "#00FFFF", "#FFA500"),guide = guide_legend(keywidth=0.6, keyheight=0.6, label.theme=element_text(size=8))) +scale_size_continuous(range=c(0.5, 3),guide = guide_legend(keywidth=0.6, keyheight=0.6, label.theme=element_text(size=8)))pdf("PCoA2_Group2_vs_Group6.pdf")p2dev.off()# ------------------------------------------# 3.5.3 Hierarchical cluster (tree) analysis#input should contain hellinger!mpse_abund %<>%mp_cal_clust(.abundance = hellinger,distmethod = "bray",hclustmethod = "average", # (UPGAE)action = "add" # action is used to control which result will be returned)mpse_abundmpse_abund %>% print(width=380, n=2)#NOTE mpse_abund contains 34 varibles, no new variable, the column bray has been new calculated!# if action = 'add', the result of hierarchical cluster will be added to the MPSE object# mp_extract_internal_attr can extract it. It is a treedata object, so it can be visualized# by ggtree.sample.clust <- mpse_abund %>% mp_extract_internal_attr(name='SampleClust')#The object contained internal attribute: PCoA ADONIS SampleClustsample.clust#--> The associated data tibble abstraction: 27 × 30library(ggtree)p <- ggtree(sample.clust) +geom_tippoint(aes(color=Group)) +geom_tiplab(as_ylab = TRUE) +ggplot2::scale_x_continuous(expand=c(0, 0.01))png("hierarchical_cluster1.png", width= 1000, height=800)pdev.off()#https://bioconductor.org/packages/release/bioc/vignettes/MicrobiotaProcess/inst/doc//MicrobiotaProcess.html# mapping = aes(x = RelRareAbundanceBySample-->Group,# y = Sample-->Group,# fill = Phyla# ),library(ggtreeExtra)library(ggplot2)# Extract relative abundance of phylaphyla.tb <- mpse_abund %>%mp_extract_abundance(taxa.class=Phylum, topn=30)# The abundance of each samples is nested, it can be flatted using the unnest of tidyr.phyla.tb %<>% tidyr::unnest(cols=RareAbundanceBySample) %>% dplyr::rename(Phyla="label")phyla.tbphyla.tb %>% print(width=380, n=10)p1 <- p +geom_fruit(data=phyla.tb,geom=geom_col,mapping = aes(x = RelRareAbundanceBySample,y = Sample,fill = Phyla),orientation = "y",#offset = 0.4,pwidth = 3,axis.params = list(axis = "x",title = "The relative abundance of phyla (%)",title.size = 4,text.size = 2,vjust = 1),grid.params = list())png("hierarchical_cluster2_Phyla.png", width = 1000, height = 800)p1dev.off()# Extract relative abundance of classesclass.tb <- mpse_abund %>%mp_extract_abundance(taxa.class = Class, topn = 30)# Flatten and rename the columnsclass.tb %<>% tidyr::unnest(cols = RareAbundanceBySample) %>% dplyr::rename(Class = "label")# View the data frameclass.tb# Create the plotp1 <- p +geom_fruit(data = class.tb,geom = geom_col,mapping = aes(x = RelRareAbundanceBySample,y = Sample,fill = Class),orientation = "y",pwidth = 3,axis.params = list(axis = "x",title = "The relative abundance of classes (%)",title.size = 4,text.size = 2,vjust = 1),grid.params = list())# Save the plot to a file #ERROR-->NEED to be DEBUGGED!png("hierarchical_cluster2_Class.png", width = 1000, height = 800)print(p1)dev.off()# -----------------------# 3.6 Biomarker discoverylibrary(ggtree)library(ggtreeExtra)library(ggplot2)library(MicrobiotaProcess)library(tidytree)library(ggstar)library(forcats)mpse_abund %>% print(width=150)#mpse_abund %<>%# mp_cal_abundance( # for each samples# .abundance = RareAbundance# ) %>%# mp_cal_abundance( # for each groups# .abundance=RareAbundance,# .group=Group# )#mpse_abundmpse_abund %<>%mp_diff_analysis(.abundance = RelRareAbundanceBySample,.group = Group,first.test.alpha = 0.01, filter.p="pvalue")# The result is stored to the taxatree or otutree slot, you can use mp_extract_tree to extract the specific slot.taxa.tree <- mpse_abund %>%mp_extract_tree(type="taxatree")taxa.tree## And the result tibble of different analysis can also be extracted## with tidytree (>=0.3.5)taxa.tree %>% select(label, nodeClass, LDAupper, LDAmean, LDAlower, Sign_Group, pvalue, fdr) %>% dplyr::filter(!is.na(fdr))taxa.tree %>% print(width=150, n=1)# Since taxa.tree is treedata object, it can be visualized by ggtree and ggtreeExtrap1 <- ggtree(taxa.tree,layout="radial",size = 0.3) +geom_point(data = td_filter(!isTip),fill="white",size=1,shape=21)# display the high light of phylum clade.p2 <- p1 +geom_hilight(data = td_filter(nodeClass == "Phylum"),mapping = aes(node = node, fill = label))# display the relative abundance of features(OTU)p3 <- p2 +ggnewscale::new_scale("fill") +geom_fruit(data = td_unnest(RareAbundanceBySample),geom = geom_star,mapping = aes(x = fct_reorder(Sample, Group, .fun=min),size = RelRareAbundanceBySample,fill = Group,subset = RelRareAbundanceBySample > 0),starshape = 13,starstroke = 0.25,offset = 0.03,pwidth = 0.4,grid.params = list(linetype=2)) +scale_size_continuous(name="Relative Abundance (%)",range = c(.5, 3)) +scale_fill_manual(values=c("#1B9E77", "#D95F02"))# display the tip labels of taxa treep4 <- p3 + geom_tiplab(size=6, offset=4.0)# display the LDA of significant OTU.#p5 <- p4 +# ggnewscale::new_scale("fill") +# geom_fruit(# geom = geom_col,# mapping = aes(# x = LDAmean,# fill = Sign_Group,# subset = !is.na(LDAmean)# ),# orientation = "y",# offset = 0.3,# pwidth = 0.5,# axis.params = list(axis = "x",# title = "Log10(LDA)",# title.height = 0.01,# title.size = 2,# text.size = 1.8,# vjust = 1),# grid.params = list(linetype = 2)# )# display the significant (FDR-->pvalue) taxonomy after kruskal.test (default)#shape = 21,#scale_size_continuous(range=c(1, 3)) +p6 <- p4 +ggnewscale::new_scale("size") +geom_point(data=td_filter(!is.na(Sign_Group)),mapping = aes(size = -log10(pvalue),fill = Sign_Group,),shape = 21,) +scale_size_continuous(range=c(1, 4)) +scale_fill_manual(values=c("#1B9E77", "#D95F02"))svg("diff_analysis_Group2_vs_Group6.svg",width=22, height=22)#png("differently_expressed_otu.png", width=2000, height=2000)p6 + theme(legend.key.height = unit(1.0, "cm"),legend.key.width = unit(1.0, "cm"),legend.spacing.y = unit(0.01, "cm"),legend.text = element_text(size = 20),legend.title = element_text(size = 20)#legend.position = c(0.99, 0.01))dev.off()#-- save the data in an Excel-file --#install.packages("tidyverse")#library(tidyverse)install.packages("dplyr")install.packages("readr")library(dplyr)library(readr)# Extracting different parts of mpse_abund, assuming these functions are applicable:abundance_data <- as.data.frame(mp_extract_abundance(mpse_abund))taxonomy_data <- as.data.frame(mp_extract_taxonomy(mpse_abund))# Joining the data together by a common identifier, assuming 'OTU' is present in bothcombined_data <- left_join(abundance_data, taxonomy_data, by = "OTU")# Install and load openxlsx package if not already installedif (!require(openxlsx)) {install.packages("openxlsx")library(openxlsx)}# Assuming `abundance_data` has lists that need flattening; example for one column:abundance_data <- abundance_data %>%mutate(RareAbundanceBySample = sapply(RareAbundanceBySample, function(x) paste(x, collapse = ", "))) %>%mutate(RareAbundanceByGroup = sapply(RareAbundanceByGroup, function(x) paste(x, collapse = ", ")))# Check the structure nowstr(abundance_data)# Rename the 'label' column in abundance_data to 'OTU'colnames(abundance_data)[colnames(abundance_data) == "label"] <- "OTU"# Merge the cleaned abundance data with taxonomy datacombined_data <- left_join(abundance_data, taxonomy_data, by = "OTU")# Check the resulting data framestr(combined_data)library(openxlsx)# Write the data frame to an Excel filewrite.xlsx(combined_data, "diff_analysis_Group2_vs_Group6.xlsx")#delete RareAbundanceBySample!# ------ Deprecated ------#.data, layout, tree.type, .taxa.class, tiplab.size, offset.abun, pwidth.abun, offset.effsize, pwidth.effsize, group.abun, tiplab.linetypep <- mpse_abund %>%mp_plot_diff_res(group.abun = FALSE,pwidth.abun=0.01,offset.abun=0.2,pwidth.effsize=0.01,offset.effsize=0.2,tiplab.size = 5.0,tiplab.linetype = 3) +scale_fill_manual(values=c("deepskyblue", "orange")) +scale_fill_manual(aesthetics = "fill_new", # The fill aes was renamed to "fill_new" for the abundance dotplot layervalues = c("deepskyblue", "orange")) +scale_fill_manual(aesthetics = "fill_new_new", # The fill aes for hight light layer of tree was renamed to 'fill_new_new'values = c("#E41A1C", "#377EB8", "#4DAF4A","#984EA3", "#FF7F00", "#FFFF33","#A65628", "#F781BF", "#00FFFF", "#999999")) +theme(axis.title = element_text(size = 28), # Font size for axis titlesaxis.text = element_text(size = 28), # Font size for axis textplot.title = element_text(size = 16), # Font size for plot titlelegend.title = element_text(size = 16), # Font size for legend titlelegend.text = element_text(size = 14) # Font size for legend text)#p$layers[[2]]$geom <- geom_tiplab(fontsize = 22) # Change 12 to the desired font sizep <- mpse_abund %>%mp_plot_diff_res(group.abun = TRUE,pwidth.abun=0.05,offset.abun=0.04,pwidth.effsize=0.3,offset.effsize=0.6,tiplab.size = 4.9,barplot.x = -log10(pvalue)) +scale_fill_manual(values=c("deepskyblue", "orange")) +scale_fill_manual(aesthetics = "fill_new", # The fill aes was renamed to "fill_new" for the abundance dotplot layervalues = c("deepskyblue", "orange")) +scale_fill_manual(aesthetics = "fill_new_new", # The fill aes for hight light layer of tree was renamed to 'fill_new_new'values = c("#E41A1C", "#377EB8", "#4DAF4A","#984EA3", "#FF7F00", "#FFFF33","#A65628", "#F781BF", "#00FFFF", "#999999")) +theme(axis.title = element_text(size = 28), # Font size for axis titlesaxis.text = element_text(size = 28), # Font size for axis textplot.title = element_text(size = 28), # Font size for plot titlelegend.title = element_text(size = 16), # Font size for legend titlelegend.text = element_text(size = 14) # Font size for legend text)#png("differently_expressed_otu.png", width=2000, height=2000)svg("p7.svg",width=22, height=22)pdev.off()p <- mpse_abund %>%mp_plot_diff_res(group.abun = FALSE,pwidth.abun=0.1,tiplab.size = 0.1,offset.effsize=0.1,pwidth.effsize=0.2) +scale_fill_manual(values=c("deepskyblue", "orange")) +scale_fill_manual(aesthetics = "fill_new", # The fill aes was renamed to "fill_new" for the abundance dotplot layervalues = c("deepskyblue", "orange")) +scale_fill_manual(aesthetics = "fill_new_new", # The fill aes for hight light layer of tree was renamed to 'fill_new_new'values = c("#E41A1C", "#377EB8", "#4DAF4A","#984EA3", "#FF7F00", "#FFFF33","#A65628", "#F781BF", "#00FFFF", "#999999"))#svg("p8.svg",width=22, height=22)png("differently_expressed_otu.png", width=2000, height=2000)pdev.off()f <- mpse_abund %>%mp_plot_diff_cladogram(label.size = 2.5,hilight.alpha = .3,bg.tree.size = .5,bg.point.size = 2,bg.point.stroke = .25) +scale_fill_diff_cladogram( # set the color of different group.values = c('deepskyblue', 'orange')) +scale_size_continuous(range = c(1, 4))#png("cladogram.png", width=1000, height=1000)svg("cladogram.svg", width=10, height=10)fdev.off()## Extract the OTU table and taxonomy table from the phyloseq objectotu_table <- phyloseq::otu_table(ps.ng.tax_abund) %>% as.data.frame() %>% as.matrix()tax_table <- phyloseq::tax_table(ps.ng.tax_abund) %>% as.data.frame() %>% as.matrix()write.csv(otu_table, file="otu_table.csv")write.csv(tax_table, file="tax_table.csv")#~/Tools/csv2xls-0.4/csv_to_xls.py otu_table.csv tax_table.csv -d',' -o otu_tax.xls
-
code of Phyloseq.Rmd
author: ""date: '`r format(Sys.time(), "%d %m %Y")`'header-includes:- \usepackage{color, fancyvrb}output:rmdformats::readthedown:highlight: katenumber_sections : yespdf_document:toc: yestoc_depth: 2number_sections : yes---```{r, echo=FALSE, warning=FALSE}## Global options# TODO: reproduce the html with the additional figure/SVN-files for editing.# IMPORTANT NOTE: needs before "mkdir figures"#rmarkdown::render('Phyloseq.Rmd',output_file='Phyloseq.html')``````{r load-packages, include=FALSE}library(knitr)library(rmdformats)library(readxl)library(dplyr)library(kableExtra)options(max.print="75")knitr::opts_chunk$set(fig.width=8,fig.height=6,eval=TRUE,cache=TRUE,echo=TRUE,prompt=FALSE,tidy=TRUE,comment=NA,message=FALSE,warning=FALSE)opts_knit$set(width=85)# Phyloseq R library#* Phyloseq web site : https://joey711.github.io/phyloseq/index.html#* See in particular tutorials for# - importing data: https://joey711.github.io/phyloseq/import-data.html# - heat maps: https://joey711.github.io/phyloseq/plot_heatmap-examples.html```# DataImport raw data and assign sample key:```{r, echo=TRUE, warning=FALSE}#extend map_corrected.txt with Diet and Flora#setwd("~/DATA/Data_Laura_16S_2/core_diversity_e4753")map_corrected <- read.csv("map_corrected.txt", sep="\t", row.names=1)knitr::kable(map_corrected) %>% kable_styling(bootstrap_options = c("striped", "hover", "condensed", "responsive"))```# Prerequisites to be installed* R : https://pbil.univ-lyon1.fr/CRAN/* R studio : https://www.rstudio.com/products/rstudio/download/#download```Rinstall.packages("dplyr") # To manipulate dataframesinstall.packages("readxl") # To read Excel files into Rinstall.packages("ggplot2") # for high quality graphicsinstall.packages("heatmaply")source("https://bioconductor.org/biocLite.R")biocLite("phyloseq")``````{r libraries, echo=TRUE, message=FALSE}library("readxl") # necessary to import the data from Excel filelibrary("ggplot2") # graphicslibrary("picante")library("microbiome") # data analysis and visualisationlibrary("phyloseq") # also the basis of data object. Data analysis and visualisationlibrary("ggpubr") # publication quality figures, based on ggplot2library("dplyr") # data handling, filter and reformat data frameslibrary("RColorBrewer") # nice color optionslibrary("heatmaply")library(vegan)library(gplots)```# Read the data and create phyloseq objectsThree tables are needed* OTU* Taxonomy* Samples```{r, echo=TRUE, warning=FALSE}#Change your working directory to where the files are locatedps.ng.tax <- import_biom("./table_even42369.biom", "./clustering/rep_set.tre")sample <- read.csv("./map_corrected.txt", sep="\t", row.names=1)SAM = sample_data(sample, errorIfNULL = T)rownames(SAM) <-c("1","2","3","5","6","7","8","9","10","12","13","14","15","16","17","18","19","20","21","22","23","24","25","26","27","28","29","30","31","32","33","34","35","36","37","38","39","40","41","42","43","44","46","47","48","49","50","51","52","53","55")ps.ng.tax <- merge_phyloseq(ps.ng.tax, SAM)print(ps.ng.tax)colnames(tax_table(ps.ng.tax)) <- c("Domain","Phylum","Class","Order","Family","Genus","Species")saveRDS(ps.ng.tax, "./ps.ng.tax.rds")```Visualize data```{r, echo=TRUE, warning=FALSE}sample_names(ps.ng.tax)rank_names(ps.ng.tax)sample_variables(ps.ng.tax)```Normalize number of reads in each sample using median sequencing depth.```{r, echo=TRUE, warning=FALSE}# RAREFACTION#set.seed(9242) # This will help in reproducing the filtering and nomalisation.#ps.ng.tax <- rarefy_even_depth(ps.ng.tax, sample.size = 42369)#total <- 42369# NORMALIZE number of reads in each sample using median sequencing depth.total = median(sample_sums(ps.ng.tax))#> total#[1] 42369standf = function(x, t=total) round(t * (x / sum(x)))ps.ng.tax = transform_sample_counts(ps.ng.tax, standf)ps.ng.tax_rel <- microbiome::transform(ps.ng.tax, "compositional")saveRDS(ps.ng.tax, "./ps.ng.tax.rds")hmp.meta <- meta(ps.ng.tax)hmp.meta$sam_name <- rownames(hmp.meta)```# Heatmaps```{r, echo=TRUE, warning=FALSE}#MOVE_FROM_ABOVE: The number of reads used for normalization is **`r sprintf("%.0f", total)`**.#A basic heatmap using the default parameters.# plot_heatmap(ps.ng.tax, method = "NMDS", distance = "bray")#NOTE that giving the correct OTU numbers in the text (1%, 0.5%, ...)!!!```We consider the most abundant OTUs for heatmaps. For example one can only take OTUs that represent at least 1% of reads in at least one sample. Remember we normalized all the sampples to median number of reads (total). We are left with only 168 OTUS which makes the reading much more easy.```{r, echo=TRUE, warning=FALSE}# Custom function to plot a heatmap with the specified sample order#plot_heatmap_custom <- function(ps, sample_order, method = "NMDS", distance = "bray") {ps.ng.tax_abund <- phyloseq::filter_taxa(ps.ng.tax, function(x) sum(x > total*0.01) > 0, TRUE)kable(otu_table(ps.ng.tax_abund)) %>%kable_styling(bootstrap_options = c("striped", "hover", "condensed", "responsive"))# Calculate the relative abundance for each sampleps.ng.tax_abund_rel <- transform_sample_counts(ps.ng.tax_abund, function(x) x / sum(x))datamat_ = as.data.frame(otu_table(ps.ng.tax_abund))#datamat <- datamat_[c("1","2","5","6","7", "8","9","10","12","13","14", "15","16","17","18","19","20", "21","22","23","24","25","26","27","28", "29","30","31","32", "33","34","35","36","37","38","39","51", "40","41","42","43","44","46", "47","48","49","50","52","53","55")]datamat <- datamat_[c("8","9","10","12","13","14", "21","22","23","24","25","26","27","28", "33","34","35","36","37","38","39","51", "47","48","49","50","52","53","55")]hr <- hclust(as.dist(1-cor(t(datamat), method="pearson")), method="complete")hc <- hclust(as.dist(1-cor(datamat, method="spearman")), method="complete")mycl = cutree(hr, h=max(hr$height)/1.08)mycol = c("YELLOW", "DARKBLUE", "DARKORANGE", "DARKMAGENTA", "DARKCYAN", "DARKRED", "MAROON", "DARKGREEN", "LIGHTBLUE", "PINK", "MAGENTA", "LIGHTCYAN","LIGHTGREEN", "BLUE", "ORANGE", "CYAN", "RED", "GREEN");mycol = mycol[as.vector(mycl)]sampleCols <- rep('GREY',ncol(datamat))#names(sampleCols) <- c("Group1", "Group1", "Group1", "Group1", "Group1", "Group2", "Group2", "Group2", "Group2", "Group2","Group2", "Group3", "Group3", "Group3", "Group3", "Group1", "Group1", "Group1", "Group1", "Group1", "Group1", "Group1", "Group1", "Group1", "Group1", "Group1", "Group1", "Group1", "Group1", "Group1", "Group1", "Group1", "Group1", "Group1", "Group1", "Group1", "Group1", "Group1", "Group1", "Group1", "Group1", "Group1", "Group1", "Group1", "Group1", "Group1", "Group1", "Group1", "Group1", "Group1")#sampleCols[colnames(datamat)=='1'] <- '#a6cee3'#sampleCols[colnames(datamat)=='2'] <- '#a6cee3'#sampleCols[colnames(datamat)=='5'] <- '#a6cee3'#sampleCols[colnames(datamat)=='6'] <- '#a6cee3'#sampleCols[colnames(datamat)=='7'] <- '#a6cee3'sampleCols[colnames(datamat)=='8'] <- '#1f78b4'sampleCols[colnames(datamat)=='9'] <- '#1f78b4'sampleCols[colnames(datamat)=='10'] <- '#1f78b4'sampleCols[colnames(datamat)=='12'] <- '#1f78b4'sampleCols[colnames(datamat)=='13'] <- '#1f78b4'sampleCols[colnames(datamat)=='14'] <- '#1f78b4'#sampleCols[colnames(datamat)=='15'] <- '#b2df8a'#sampleCols[colnames(datamat)=='16'] <- '#b2df8a'#sampleCols[colnames(datamat)=='17'] <- '#b2df8a'#sampleCols[colnames(datamat)=='18'] <- '#b2df8a'#sampleCols[colnames(datamat)=='19'] <- '#b2df8a'#sampleCols[colnames(datamat)=='20'] <- '#b2df8a'sampleCols[colnames(datamat)=='21'] <- '#33a02c'sampleCols[colnames(datamat)=='22'] <- '#33a02c'sampleCols[colnames(datamat)=='23'] <- '#33a02c'sampleCols[colnames(datamat)=='24'] <- '#33a02c'sampleCols[colnames(datamat)=='25'] <- '#33a02c'sampleCols[colnames(datamat)=='26'] <- '#33a02c'sampleCols[colnames(datamat)=='27'] <- '#33a02c'sampleCols[colnames(datamat)=='28'] <- '#33a02c'#sampleCols[colnames(datamat)=='29'] <- '#fb9a99'#sampleCols[colnames(datamat)=='30'] <- '#fb9a99'#sampleCols[colnames(datamat)=='31'] <- '#fb9a99'#sampleCols[colnames(datamat)=='32'] <- '#fb9a99'sampleCols[colnames(datamat)=='33'] <- '#e31a1c'sampleCols[colnames(datamat)=='34'] <- '#e31a1c'sampleCols[colnames(datamat)=='35'] <- '#e31a1c'sampleCols[colnames(datamat)=='36'] <- '#e31a1c'sampleCols[colnames(datamat)=='37'] <- '#e31a1c'sampleCols[colnames(datamat)=='38'] <- '#e31a1c'sampleCols[colnames(datamat)=='39'] <- '#e31a1c'sampleCols[colnames(datamat)=='51'] <- '#e31a1c'#sampleCols[colnames(datamat)=='40'] <- '#cab2d6'#sampleCols[colnames(datamat)=='41'] <- '#cab2d6'#sampleCols[colnames(datamat)=='42'] <- '#cab2d6'#sampleCols[colnames(datamat)=='43'] <- '#cab2d6'#sampleCols[colnames(datamat)=='44'] <- '#cab2d6'#sampleCols[colnames(datamat)=='46'] <- '#cab2d6'sampleCols[colnames(datamat)=='47'] <- '#6a3d9a'sampleCols[colnames(datamat)=='48'] <- '#6a3d9a'sampleCols[colnames(datamat)=='49'] <- '#6a3d9a'sampleCols[colnames(datamat)=='50'] <- '#6a3d9a'sampleCols[colnames(datamat)=='52'] <- '#6a3d9a'sampleCols[colnames(datamat)=='53'] <- '#6a3d9a'sampleCols[colnames(datamat)=='55'] <- '#6a3d9a'#bluered(75)#color_pattern <- colorRampPalette(c("blue", "white", "red"))(100)library(RColorBrewer)custom_palette <- colorRampPalette(brewer.pal(9, "Blues"))heatmap_colors <- custom_palette(100)#colors <- heatmap_color_default(100)png("figures/heatmap.png", width=1200, height=2400)#par(mar=c(2, 2, 2, 2)) , lwid=1 lhei=c(0.7, 10)) # Adjust height of color keys keysize=0.3,heatmap.2(as.matrix(datamat),Rowv=as.dendrogram(hr),Colv = NA, dendrogram = 'row',scale='row',trace='none',col=heatmap_colors, cexRow=1.2, cexCol=1.5,RowSideColors = mycol, ColSideColors = sampleCols, srtCol=15, labRow=row.names(datamat), key=TRUE, margins=c(10, 15), lhei=c(0.7, 15), lwid=c(1,8))dev.off()``````{r, echo=TRUE, warning=FALSE, fig.cap="Heatmap", out.width = '100%', fig.align= "center"}knitr::include_graphics("./figures/heatmap.png")``````{r, echo=FALSE, warning=FALSE}#It is possible to use different distances and different multivaraite methods. Many different built-in distances can be used.#dist_methods <- unlist(distanceMethodList)#print(dist_methods)```\pagebreak# Taxonomic summary## Bar plots in phylum level```{r, echo=FALSE, warning=FALSE}#Make the bargraph nicer by removing OTUs boundaries. This is done by adding ggplot2 modifier.# 1: uniform color. Color is for the border, fill is for the inside#ggplot(mtcars, aes(x=as.factor(cyl) )) +# geom_bar(color="blue", fill=rgb(0.1,0.4,0.5,0.7) )# 2: Using Hue#ggplot(mtcars, aes(x=as.factor(cyl), fill=as.factor(cyl) )) +# geom_bar( ) +# scale_fill_hue(c = 40) +# theme(legend.position="none")# 3: Using RColorBrewer#ggplot(mtcars, aes(x=as.factor(cyl), fill=as.factor(cyl) )) +# geom_bar( ) +# scale_fill_brewer(palette = "Set1") +# theme(legend.position="none")# 4: Using greyscale:#ggplot(mtcars, aes(x=as.factor(cyl), fill=as.factor(cyl) )) +# geom_bar( ) +# scale_fill_grey(start = 0.25, end = 0.75) +# theme(legend.position="none")# 5: Set manualy#ggplot(mtcars, aes(x=as.factor(cyl), fill=as.factor(cyl) )) +# geom_bar( ) +# scale_fill_manual(values = c("red", "green", "blue") ) +# theme(legend.position="none")#NOT SUCCESSFUL!#allGroupsColors<- c(# "grey0", "grey50", "dodgerblu", "deepskyblue",# "red", "darkred", "green", "green4")# plot_bar(ps.ng.tax_rel, fill="Phylum") +# geom_bar(stat="identity", position="stack") + scale_color_manual(values = allGroupsColors) #, fill=Phylum + scale_fill_brewer(palette = "Set1")# ##### Keep only the most abundant phyla and#ps.ng.tax_most <- subset_taxa(ps.ng.tax_rel, Phylum %in% c("D_1__Actinobacteria")) #1.57#ps.ng.tax_most <- subset_taxa(ps.ng.tax_rel, Phylum %in% c("D_1__Bacteroidetes")) #27.27436#ps.ng.tax_most <- subset_taxa(ps.ng.tax_rel, Phylum %in% c("D_1__Cyanobacteria")) #0.02244249#ps.ng.tax_most <- subset_taxa(ps.ng.tax_rel, Phylum %in% c("D_1__Epsilonbacteraeota")) #0.01309145#ps.ng.tax_most <- subset_taxa(ps.ng.tax_rel, Phylum %in% c("D_1__Euryarchaeota")) #0.1210024#ps.ng.tax_most <- subset_taxa(ps.ng.tax_rel, Phylum %in% c("D_1__Firmicutes")) #32.50589#ps.ng.tax_most <- subset_taxa(ps.ng.tax_rel, Phylum %in% c("D_1__Lentisphaerae")) #0.0001870208#ps.ng.tax_most <- subset_taxa(ps.ng.tax_rel, Phylum %in% c("D_1__Patescibacteria")) #0.008789976#ps.ng.tax_most <- subset_taxa(ps.ng.tax_rel, Phylum %in% c("D_1__Planctomycetes")) #0.01365252#ps.ng.tax_most <- subset_taxa(ps.ng.tax_rel, Phylum %in% c("D_1__Proteobacteria")) #6.769216#ps.ng.tax_most <- subset_taxa(ps.ng.tax_rel, Phylum %in% c("D_1__Synergistetes")) #0.005049561#ps.ng.tax_most <- subset_taxa(ps.ng.tax_rel, Phylum %in% c("D_1__Tenericutes")) #0.0005610623#ps.ng.tax_most <- subset_taxa(ps.ng.tax_rel, Phylum %in% c("D_1__Verrucomicrobia")) #2.076304#ps.ng.tax_most <- subset_taxa(ps.ng.tax_rel, Phylum %in% c(NA)) #sum(otu_table(ps.ng.tax_most)) = 2.619413``````{r, echo=TRUE, warning=FALSE}library(ggplot2)geom.text.size = 6theme.size = 8 #(14/5) * geom.text.size#ps.ng.tax_most <- subset_taxa(ps.ng.tax_rel, Phylum %in% c("D_1__Actinobacteria", "D_1__Bacteroidetes", "D_1__Firmicutes", "D_1__Proteobacteria", "D_1__Verrucomicrobia", NA))ps.ng.tax_most = phyloseq::filter_taxa(ps.ng.tax_rel, function(x) mean(x) > 0.001, TRUE)#CONSOLE(OPTIONAL): for sampleid in 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73; do#echo "otu_table(ps.ng.tax_most)[,${sampleid}]=otu_table(ps.ng.tax_most)[,${sampleid}]/sum(otu_table(ps.ng.tax_most)[,${sampleid}])" done#ORps.ng.tax_most_ = transform_sample_counts(ps.ng.tax_most, function(x) x / sum(x))``````{r, echo=FALSE, warning=FALSE}##--Creating 100% stacked bar plots with less abundant taxa in a sub-category #901--##https://github.com/joey711/phyloseq/issues/901##ps.ng.tax_most_df <- psmelt(ps.ng.tax_most_) #5986x19#glom <- tax_glom(ps.ng.tax_most_, taxrank = 'Phylum')#tax_table(glom) # should list # taxa as # phyla#data <- psmelt(glom) # create dataframe from phyloseq object#data$Phylum <- as.character(data$Phylum) #convert to character##simple way to rename phyla with < 1% abundance#data$Phylum[data$Abundance < 0.001] <- "< 0.1% abund."##library(plyr)#medians <- ddply(data, ~Phylum, function(x) c(median=median(x$Abundance)))#remainder <- medians[medians$median <= 0.001,]$Phylum#data[data$Phylum %in% remainder,]$Phylum <- "Phyla < 0.1% abund."#data$Phylum[data$Abundance < 0.001] <- "Phyla < 0.1% abund."##--> data are not used!###in class level#glom <- tax_glom(ps.ng.tax_most_, taxrank = 'Class')#tax_table(glom) # should list # taxa as # phyla#data <- psmelt(glom) # create dataframe from phyloseq object#data$Class <- as.character(data$Class) #convert to character###simple way to rename phyla with < 1% abundance#data$Class[data$Abundance < 0.001] <- "< 0.1% abund."#Count = length(unique(data$Class))###unique(data$Class)##data$Class <- factor(data$Class, levels = c("Bacilli", "Bacteroidia", "Verrucomicrobiae", "Clostridia", "Gammaproteobacteria", "Alphaproteobacteria", "Actinobacteria", "Negativicutes", "Erysipelotrichia", "Methanobacteria", "< 0.1% abund."))##------- Creating 100% stacked bar plots END --------library(stringr)#FITTING1:# tax_table(ps.ng.tax_most_)[1,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[1,"Domain"], "__")[[1]][2]# ... ...# tax_table(ps.ng.tax_most_)[167,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[167,"Species"], "__")[[1]][2]#ps.ng.tax_most_#in total [ 89 taxa and 55 samples ]#otu_table() OTU Table: [ 166 taxa and 54 samples ]#otu_table() OTU Table: [ 168 taxa and 50 samples ]#for id in 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112 113 114 115 116 117 118 119 120 121 122 123 124 125 126 127 128 129 130 131 132 133 134 135 136 137 138 139 140 141 142 143 144 145 146 147 148 149 150 151 152 153 154 155 156 157 158 159 160 161 162 163 164 165 166; do#echo "tax_table(ps.ng.tax_most_)[${id},\"Domain\"] <- str_split(tax_table(ps.ng.tax_most_)[${id},\"Domain\"], \"__\")[[1]][2]"#echo "tax_table(ps.ng.tax_most_)[${id},\"Phylum\"] <- str_split(tax_table(ps.ng.tax_most_)[${id},\"Phylum\"], \"__\")[[1]][2]"#echo "tax_table(ps.ng.tax_most_)[${id},\"Class\"] <- str_split(tax_table(ps.ng.tax_most_)[${id},\"Class\"], \"__\")[[1]][2]"#echo "tax_table(ps.ng.tax_most_)[${id},\"Order\"] <- str_split(tax_table(ps.ng.tax_most_)[${id},\"Order\"], \"__\")[[1]][2]"#echo "tax_table(ps.ng.tax_most_)[${id},\"Family\"] <- str_split(tax_table(ps.ng.tax_most_)[${id},\"Family\"], \"__\")[[1]][2]"#echo "tax_table(ps.ng.tax_most_)[${id},\"Genus\"] <- str_split(tax_table(ps.ng.tax_most_)[${id},\"Genus\"], \"__\")[[1]][2]"#echo "tax_table(ps.ng.tax_most_)[${id},\"Species\"] <- str_split(tax_table(ps.ng.tax_most_)[${id},\"Species\"], \"__\")[[1]][2]"#donetax_table(ps.ng.tax_most_)[1,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[1,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[1,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[1,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[1,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[1,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[1,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[1,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[1,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[1,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[1,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[1,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[1,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[1,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[2,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[2,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[2,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[2,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[2,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[2,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[2,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[2,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[2,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[2,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[2,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[2,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[2,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[2,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[3,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[3,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[3,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[3,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[3,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[3,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[3,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[3,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[3,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[3,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[3,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[3,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[3,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[3,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[4,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[4,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[4,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[4,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[4,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[4,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[4,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[4,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[4,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[4,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[4,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[4,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[4,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[4,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[5,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[5,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[5,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[5,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[5,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[5,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[5,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[5,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[5,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[5,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[5,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[5,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[5,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[5,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[6,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[6,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[6,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[6,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[6,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[6,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[6,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[6,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[6,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[6,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[6,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[6,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[6,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[6,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[7,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[7,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[7,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[7,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[7,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[7,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[7,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[7,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[7,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[7,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[7,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[7,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[7,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[7,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[8,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[8,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[8,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[8,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[8,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[8,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[8,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[8,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[8,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[8,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[8,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[8,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[8,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[8,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[9,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[9,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[9,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[9,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[9,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[9,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[9,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[9,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[9,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[9,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[9,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[9,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[9,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[9,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[10,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[10,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[10,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[10,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[10,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[10,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[10,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[10,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[10,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[10,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[10,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[10,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[10,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[10,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[11,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[11,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[11,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[11,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[11,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[11,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[11,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[11,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[11,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[11,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[11,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[11,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[11,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[11,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[12,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[12,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[12,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[12,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[12,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[12,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[12,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[12,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[12,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[12,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[12,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[12,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[12,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[12,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[13,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[13,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[13,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[13,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[13,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[13,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[13,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[13,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[13,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[13,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[13,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[13,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[13,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[13,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[14,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[14,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[14,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[14,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[14,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[14,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[14,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[14,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[14,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[14,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[14,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[14,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[14,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[14,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[15,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[15,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[15,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[15,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[15,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[15,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[15,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[15,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[15,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[15,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[15,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[15,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[15,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[15,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[16,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[16,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[16,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[16,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[16,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[16,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[16,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[16,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[16,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[16,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[16,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[16,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[16,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[16,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[17,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[17,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[17,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[17,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[17,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[17,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[17,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[17,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[17,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[17,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[17,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[17,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[17,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[17,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[18,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[18,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[18,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[18,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[18,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[18,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[18,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[18,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[18,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[18,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[18,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[18,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[18,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[18,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[19,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[19,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[19,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[19,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[19,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[19,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[19,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[19,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[19,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[19,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[19,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[19,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[19,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[19,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[20,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[20,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[20,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[20,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[20,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[20,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[20,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[20,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[20,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[20,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[20,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[20,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[20,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[20,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[21,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[21,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[21,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[21,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[21,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[21,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[21,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[21,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[21,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[21,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[21,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[21,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[21,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[21,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[22,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[22,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[22,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[22,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[22,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[22,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[22,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[22,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[22,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[22,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[22,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[22,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[22,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[22,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[23,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[23,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[23,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[23,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[23,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[23,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[23,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[23,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[23,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[23,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[23,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[23,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[23,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[23,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[24,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[24,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[24,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[24,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[24,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[24,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[24,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[24,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[24,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[24,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[24,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[24,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[24,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[24,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[25,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[25,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[25,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[25,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[25,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[25,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[25,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[25,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[25,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[25,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[25,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[25,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[25,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[25,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[26,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[26,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[26,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[26,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[26,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[26,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[26,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[26,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[26,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[26,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[26,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[26,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[26,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[26,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[27,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[27,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[27,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[27,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[27,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[27,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[27,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[27,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[27,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[27,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[27,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[27,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[27,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[27,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[28,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[28,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[28,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[28,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[28,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[28,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[28,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[28,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[28,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[28,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[28,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[28,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[28,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[28,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[29,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[29,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[29,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[29,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[29,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[29,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[29,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[29,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[29,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[29,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[29,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[29,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[29,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[29,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[30,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[30,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[30,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[30,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[30,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[30,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[30,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[30,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[30,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[30,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[30,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[30,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[30,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[30,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[31,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[31,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[31,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[31,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[31,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[31,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[31,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[31,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[31,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[31,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[31,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[31,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[31,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[31,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[32,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[32,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[32,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[32,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[32,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[32,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[32,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[32,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[32,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[32,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[32,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[32,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[32,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[32,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[33,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[33,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[33,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[33,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[33,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[33,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[33,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[33,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[33,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[33,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[33,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[33,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[33,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[33,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[34,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[34,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[34,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[34,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[34,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[34,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[34,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[34,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[34,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[34,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[34,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[34,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[34,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[34,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[35,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[35,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[35,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[35,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[35,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[35,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[35,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[35,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[35,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[35,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[35,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[35,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[35,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[35,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[36,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[36,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[36,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[36,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[36,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[36,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[36,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[36,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[36,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[36,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[36,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[36,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[36,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[36,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[37,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[37,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[37,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[37,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[37,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[37,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[37,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[37,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[37,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[37,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[37,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[37,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[37,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[37,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[38,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[38,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[38,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[38,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[38,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[38,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[38,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[38,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[38,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[38,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[38,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[38,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[38,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[38,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[39,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[39,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[39,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[39,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[39,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[39,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[39,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[39,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[39,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[39,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[39,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[39,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[39,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[39,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[40,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[40,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[40,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[40,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[40,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[40,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[40,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[40,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[40,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[40,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[40,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[40,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[40,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[40,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[41,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[41,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[41,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[41,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[41,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[41,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[41,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[41,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[41,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[41,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[41,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[41,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[41,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[41,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[42,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[42,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[42,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[42,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[42,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[42,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[42,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[42,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[42,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[42,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[42,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[42,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[42,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[42,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[43,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[43,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[43,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[43,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[43,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[43,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[43,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[43,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[43,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[43,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[43,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[43,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[43,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[43,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[44,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[44,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[44,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[44,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[44,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[44,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[44,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[44,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[44,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[44,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[44,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[44,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[44,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[44,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[45,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[45,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[45,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[45,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[45,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[45,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[45,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[45,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[45,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[45,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[45,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[45,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[45,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[45,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[46,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[46,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[46,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[46,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[46,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[46,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[46,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[46,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[46,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[46,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[46,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[46,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[46,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[46,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[47,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[47,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[47,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[47,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[47,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[47,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[47,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[47,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[47,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[47,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[47,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[47,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[47,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[47,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[48,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[48,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[48,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[48,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[48,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[48,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[48,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[48,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[48,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[48,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[48,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[48,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[48,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[48,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[49,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[49,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[49,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[49,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[49,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[49,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[49,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[49,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[49,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[49,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[49,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[49,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[49,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[49,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[50,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[50,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[50,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[50,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[50,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[50,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[50,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[50,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[50,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[50,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[50,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[50,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[50,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[50,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[51,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[51,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[51,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[51,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[51,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[51,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[51,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[51,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[51,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[51,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[51,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[51,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[51,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[51,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[52,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[52,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[52,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[52,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[52,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[52,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[52,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[52,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[52,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[52,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[52,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[52,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[52,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[52,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[53,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[53,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[53,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[53,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[53,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[53,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[53,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[53,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[53,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[53,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[53,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[53,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[53,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[53,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[54,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[54,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[54,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[54,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[54,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[54,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[54,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[54,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[54,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[54,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[54,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[54,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[54,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[54,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[55,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[55,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[55,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[55,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[55,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[55,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[55,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[55,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[55,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[55,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[55,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[55,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[55,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[55,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[56,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[56,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[56,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[56,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[56,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[56,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[56,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[56,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[56,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[56,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[56,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[56,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[56,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[56,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[57,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[57,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[57,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[57,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[57,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[57,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[57,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[57,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[57,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[57,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[57,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[57,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[57,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[57,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[58,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[58,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[58,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[58,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[58,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[58,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[58,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[58,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[58,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[58,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[58,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[58,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[58,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[58,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[59,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[59,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[59,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[59,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[59,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[59,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[59,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[59,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[59,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[59,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[59,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[59,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[59,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[59,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[60,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[60,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[60,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[60,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[60,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[60,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[60,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[60,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[60,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[60,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[60,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[60,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[60,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[60,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[61,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[61,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[61,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[61,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[61,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[61,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[61,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[61,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[61,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[61,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[61,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[61,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[61,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[61,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[62,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[62,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[62,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[62,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[62,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[62,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[62,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[62,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[62,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[62,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[62,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[62,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[62,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[62,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[63,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[63,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[63,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[63,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[63,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[63,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[63,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[63,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[63,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[63,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[63,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[63,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[63,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[63,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[64,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[64,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[64,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[64,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[64,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[64,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[64,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[64,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[64,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[64,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[64,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[64,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[64,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[64,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[65,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[65,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[65,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[65,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[65,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[65,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[65,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[65,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[65,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[65,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[65,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[65,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[65,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[65,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[66,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[66,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[66,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[66,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[66,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[66,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[66,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[66,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[66,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[66,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[66,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[66,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[66,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[66,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[67,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[67,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[67,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[67,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[67,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[67,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[67,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[67,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[67,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[67,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[67,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[67,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[67,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[67,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[68,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[68,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[68,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[68,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[68,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[68,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[68,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[68,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[68,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[68,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[68,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[68,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[68,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[68,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[69,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[69,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[69,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[69,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[69,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[69,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[69,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[69,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[69,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[69,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[69,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[69,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[69,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[69,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[70,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[70,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[70,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[70,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[70,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[70,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[70,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[70,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[70,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[70,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[70,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[70,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[70,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[70,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[71,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[71,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[71,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[71,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[71,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[71,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[71,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[71,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[71,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[71,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[71,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[71,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[71,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[71,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[72,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[72,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[72,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[72,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[72,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[72,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[72,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[72,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[72,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[72,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[72,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[72,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[72,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[72,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[73,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[73,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[73,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[73,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[73,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[73,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[73,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[73,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[73,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[73,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[73,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[73,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[73,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[73,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[74,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[74,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[74,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[74,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[74,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[74,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[74,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[74,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[74,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[74,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[74,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[74,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[74,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[74,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[75,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[75,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[75,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[75,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[75,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[75,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[75,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[75,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[75,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[75,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[75,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[75,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[75,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[75,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[76,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[76,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[76,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[76,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[76,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[76,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[76,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[76,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[76,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[76,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[76,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[76,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[76,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[76,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[77,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[77,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[77,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[77,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[77,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[77,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[77,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[77,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[77,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[77,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[77,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[77,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[77,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[77,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[78,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[78,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[78,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[78,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[78,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[78,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[78,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[78,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[78,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[78,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[78,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[78,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[78,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[78,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[79,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[79,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[79,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[79,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[79,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[79,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[79,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[79,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[79,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[79,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[79,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[79,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[79,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[79,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[80,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[80,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[80,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[80,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[80,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[80,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[80,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[80,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[80,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[80,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[80,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[80,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[80,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[80,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[81,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[81,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[81,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[81,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[81,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[81,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[81,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[81,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[81,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[81,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[81,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[81,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[81,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[81,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[82,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[82,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[82,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[82,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[82,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[82,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[82,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[82,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[82,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[82,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[82,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[82,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[82,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[82,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[83,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[83,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[83,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[83,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[83,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[83,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[83,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[83,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[83,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[83,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[83,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[83,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[83,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[83,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[84,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[84,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[84,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[84,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[84,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[84,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[84,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[84,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[84,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[84,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[84,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[84,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[84,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[84,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[85,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[85,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[85,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[85,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[85,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[85,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[85,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[85,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[85,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[85,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[85,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[85,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[85,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[85,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[86,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[86,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[86,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[86,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[86,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[86,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[86,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[86,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[86,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[86,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[86,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[86,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[86,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[86,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[87,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[87,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[87,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[87,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[87,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[87,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[87,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[87,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[87,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[87,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[87,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[87,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[87,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[87,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[88,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[88,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[88,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[88,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[88,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[88,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[88,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[88,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[88,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[88,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[88,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[88,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[88,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[88,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[89,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[89,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[89,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[89,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[89,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[89,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[89,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[89,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[89,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[89,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[89,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[89,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[89,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[89,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[90,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[90,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[90,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[90,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[90,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[90,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[90,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[90,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[90,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[90,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[90,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[90,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[90,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[90,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[91,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[91,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[91,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[91,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[91,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[91,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[91,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[91,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[91,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[91,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[91,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[91,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[91,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[91,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[92,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[92,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[92,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[92,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[92,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[92,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[92,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[92,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[92,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[92,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[92,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[92,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[92,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[92,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[93,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[93,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[93,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[93,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[93,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[93,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[93,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[93,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[93,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[93,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[93,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[93,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[93,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[93,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[94,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[94,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[94,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[94,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[94,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[94,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[94,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[94,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[94,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[94,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[94,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[94,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[94,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[94,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[95,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[95,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[95,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[95,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[95,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[95,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[95,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[95,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[95,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[95,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[95,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[95,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[95,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[95,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[96,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[96,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[96,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[96,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[96,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[96,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[96,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[96,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[96,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[96,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[96,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[96,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[96,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[96,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[97,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[97,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[97,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[97,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[97,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[97,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[97,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[97,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[97,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[97,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[97,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[97,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[97,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[97,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[98,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[98,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[98,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[98,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[98,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[98,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[98,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[98,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[98,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[98,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[98,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[98,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[98,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[98,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[99,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[99,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[99,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[99,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[99,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[99,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[99,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[99,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[99,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[99,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[99,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[99,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[99,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[99,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[100,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[100,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[100,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[100,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[100,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[100,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[100,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[100,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[100,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[100,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[100,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[100,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[100,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[100,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[101,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[101,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[101,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[101,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[101,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[101,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[101,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[101,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[101,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[101,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[101,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[101,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[101,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[101,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[102,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[102,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[102,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[102,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[102,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[102,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[102,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[102,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[102,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[102,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[102,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[102,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[102,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[102,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[103,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[103,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[103,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[103,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[103,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[103,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[103,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[103,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[103,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[103,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[103,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[103,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[103,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[103,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[104,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[104,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[104,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[104,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[104,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[104,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[104,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[104,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[104,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[104,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[104,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[104,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[104,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[104,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[105,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[105,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[105,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[105,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[105,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[105,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[105,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[105,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[105,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[105,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[105,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[105,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[105,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[105,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[106,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[106,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[106,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[106,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[106,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[106,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[106,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[106,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[106,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[106,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[106,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[106,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[106,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[106,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[107,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[107,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[107,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[107,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[107,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[107,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[107,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[107,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[107,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[107,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[107,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[107,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[107,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[107,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[108,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[108,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[108,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[108,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[108,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[108,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[108,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[108,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[108,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[108,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[108,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[108,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[108,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[108,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[109,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[109,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[109,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[109,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[109,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[109,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[109,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[109,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[109,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[109,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[109,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[109,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[109,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[109,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[110,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[110,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[110,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[110,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[110,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[110,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[110,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[110,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[110,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[110,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[110,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[110,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[110,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[110,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[111,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[111,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[111,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[111,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[111,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[111,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[111,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[111,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[111,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[111,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[111,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[111,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[111,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[111,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[112,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[112,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[112,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[112,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[112,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[112,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[112,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[112,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[112,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[112,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[112,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[112,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[112,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[112,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[113,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[113,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[113,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[113,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[113,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[113,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[113,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[113,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[113,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[113,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[113,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[113,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[113,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[113,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[114,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[114,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[114,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[114,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[114,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[114,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[114,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[114,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[114,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[114,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[114,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[114,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[114,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[114,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[115,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[115,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[115,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[115,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[115,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[115,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[115,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[115,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[115,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[115,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[115,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[115,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[115,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[115,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[116,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[116,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[116,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[116,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[116,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[116,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[116,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[116,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[116,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[116,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[116,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[116,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[116,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[116,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[117,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[117,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[117,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[117,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[117,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[117,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[117,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[117,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[117,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[117,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[117,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[117,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[117,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[117,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[118,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[118,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[118,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[118,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[118,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[118,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[118,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[118,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[118,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[118,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[118,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[118,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[118,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[118,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[119,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[119,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[119,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[119,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[119,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[119,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[119,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[119,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[119,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[119,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[119,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[119,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[119,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[119,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[120,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[120,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[120,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[120,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[120,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[120,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[120,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[120,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[120,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[120,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[120,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[120,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[120,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[120,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[121,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[121,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[121,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[121,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[121,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[121,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[121,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[121,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[121,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[121,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[121,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[121,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[121,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[121,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[122,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[122,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[122,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[122,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[122,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[122,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[122,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[122,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[122,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[122,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[122,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[122,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[122,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[122,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[123,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[123,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[123,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[123,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[123,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[123,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[123,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[123,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[123,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[123,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[123,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[123,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[123,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[123,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[124,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[124,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[124,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[124,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[124,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[124,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[124,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[124,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[124,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[124,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[124,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[124,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[124,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[124,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[125,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[125,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[125,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[125,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[125,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[125,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[125,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[125,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[125,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[125,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[125,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[125,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[125,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[125,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[126,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[126,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[126,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[126,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[126,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[126,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[126,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[126,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[126,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[126,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[126,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[126,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[126,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[126,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[127,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[127,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[127,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[127,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[127,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[127,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[127,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[127,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[127,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[127,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[127,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[127,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[127,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[127,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[128,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[128,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[128,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[128,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[128,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[128,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[128,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[128,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[128,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[128,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[128,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[128,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[128,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[128,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[129,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[129,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[129,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[129,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[129,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[129,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[129,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[129,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[129,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[129,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[129,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[129,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[129,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[129,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[130,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[130,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[130,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[130,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[130,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[130,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[130,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[130,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[130,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[130,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[130,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[130,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[130,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[130,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[131,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[131,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[131,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[131,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[131,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[131,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[131,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[131,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[131,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[131,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[131,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[131,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[131,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[131,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[132,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[132,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[132,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[132,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[132,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[132,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[132,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[132,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[132,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[132,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[132,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[132,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[132,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[132,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[133,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[133,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[133,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[133,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[133,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[133,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[133,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[133,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[133,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[133,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[133,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[133,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[133,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[133,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[134,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[134,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[134,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[134,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[134,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[134,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[134,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[134,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[134,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[134,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[134,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[134,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[134,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[134,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[135,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[135,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[135,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[135,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[135,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[135,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[135,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[135,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[135,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[135,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[135,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[135,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[135,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[135,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[136,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[136,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[136,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[136,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[136,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[136,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[136,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[136,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[136,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[136,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[136,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[136,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[136,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[136,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[137,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[137,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[137,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[137,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[137,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[137,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[137,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[137,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[137,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[137,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[137,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[137,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[137,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[137,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[138,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[138,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[138,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[138,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[138,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[138,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[138,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[138,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[138,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[138,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[138,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[138,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[138,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[138,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[139,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[139,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[139,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[139,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[139,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[139,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[139,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[139,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[139,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[139,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[139,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[139,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[139,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[139,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[140,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[140,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[140,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[140,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[140,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[140,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[140,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[140,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[140,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[140,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[140,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[140,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[140,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[140,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[141,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[141,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[141,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[141,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[141,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[141,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[141,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[141,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[141,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[141,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[141,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[141,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[141,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[141,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[142,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[142,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[142,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[142,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[142,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[142,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[142,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[142,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[142,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[142,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[142,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[142,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[142,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[142,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[143,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[143,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[143,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[143,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[143,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[143,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[143,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[143,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[143,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[143,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[143,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[143,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[143,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[143,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[144,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[144,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[144,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[144,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[144,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[144,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[144,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[144,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[144,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[144,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[144,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[144,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[144,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[144,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[145,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[145,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[145,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[145,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[145,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[145,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[145,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[145,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[145,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[145,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[145,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[145,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[145,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[145,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[146,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[146,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[146,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[146,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[146,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[146,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[146,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[146,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[146,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[146,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[146,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[146,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[146,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[146,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[147,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[147,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[147,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[147,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[147,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[147,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[147,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[147,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[147,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[147,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[147,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[147,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[147,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[147,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[148,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[148,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[148,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[148,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[148,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[148,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[148,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[148,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[148,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[148,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[148,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[148,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[148,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[148,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[149,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[149,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[149,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[149,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[149,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[149,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[149,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[149,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[149,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[149,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[149,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[149,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[149,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[149,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[150,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[150,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[150,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[150,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[150,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[150,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[150,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[150,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[150,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[150,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[150,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[150,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[150,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[150,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[151,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[151,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[151,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[151,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[151,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[151,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[151,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[151,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[151,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[151,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[151,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[151,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[151,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[151,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[152,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[152,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[152,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[152,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[152,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[152,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[152,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[152,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[152,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[152,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[152,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[152,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[152,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[152,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[153,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[153,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[153,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[153,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[153,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[153,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[153,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[153,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[153,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[153,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[153,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[153,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[153,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[153,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[154,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[154,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[154,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[154,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[154,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[154,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[154,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[154,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[154,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[154,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[154,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[154,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[154,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[154,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[155,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[155,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[155,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[155,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[155,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[155,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[155,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[155,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[155,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[155,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[155,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[155,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[155,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[155,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[156,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[156,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[156,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[156,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[156,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[156,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[156,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[156,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[156,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[156,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[156,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[156,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[156,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[156,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[157,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[157,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[157,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[157,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[157,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[157,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[157,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[157,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[157,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[157,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[157,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[157,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[157,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[157,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[158,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[158,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[158,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[158,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[158,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[158,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[158,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[158,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[158,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[158,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[158,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[158,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[158,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[158,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[159,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[159,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[159,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[159,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[159,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[159,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[159,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[159,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[159,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[159,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[159,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[159,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[159,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[159,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[160,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[160,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[160,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[160,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[160,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[160,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[160,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[160,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[160,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[160,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[160,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[160,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[160,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[160,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[161,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[161,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[161,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[161,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[161,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[161,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[161,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[161,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[161,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[161,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[161,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[161,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[161,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[161,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[162,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[162,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[162,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[162,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[162,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[162,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[162,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[162,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[162,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[162,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[162,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[162,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[162,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[162,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[163,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[163,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[163,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[163,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[163,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[163,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[163,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[163,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[163,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[163,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[163,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[163,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[163,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[163,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[164,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[164,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[164,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[164,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[164,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[164,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[164,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[164,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[164,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[164,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[164,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[164,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[164,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[164,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[165,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[165,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[165,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[165,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[165,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[165,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[165,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[165,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[165,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[165,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[165,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[165,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[165,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[165,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[166,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[166,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[166,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[166,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[166,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[166,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[166,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[166,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[166,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[166,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[166,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[166,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[166,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[166,"Species"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[167,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[167,"Domain"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[167,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[167,"Phylum"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[167,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[167,"Class"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[167,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[167,"Order"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[167,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[167,"Family"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[167,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[167,"Genus"], "__")[[1]][2]tax_table(ps.ng.tax_most_)[167,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[167,"Species"], "__")[[1]][2]``````{r, echo=TRUE, warning=FALSE}#aes(color="Phylum", fill="Phylum") --> aes()#ggplot(data=data, aes(x=Sample, y=Abundance, fill=Phylum))plot_bar(ps.ng.tax_most_, fill="Phylum") + geom_bar(aes(), stat="identity", position="stack") +scale_fill_manual(values = c("darkblue", "darkgoldenrod1", "darkseagreen", "darkorchid", "darkolivegreen1", "lightskyblue", "darkgreen", "deeppink", "khaki2", "firebrick", "brown1", "darkorange1", "cyan1", "royalblue4", "darksalmon", "darkblue","royalblue4", "dodgerblue3", "steelblue1", "lightskyblue", "darkseagreen", "darkgoldenrod1", "darkseagreen", "darkorchid", "darkolivegreen1", "brown1", "darkorange1", "cyan1", "darkgrey")) + theme(axis.text = element_text(size = 5, colour="black")) + theme(legend.position="bottom") + guides(fill=guide_legend(nrow=2)) #6 instead of theme.size``````{r, echo=FALSE, warning=FALSE}#png("abc.png")#knitr::include_graphics("./Phyloseq_files/figure-html/unnamed-chunk-7-1.png")#dev.off()```\pagebreakRegroup together pre vs post stroke samples and normalize number of reads in each group using median sequencing depth.```{r, echo=TRUE, warning=FALSE}ps.ng.tax_most_pre_post_stroke <- merge_samples(ps.ng.tax_most_, "pre_post_stroke")ps.ng.tax_most_pre_post_stroke_ = transform_sample_counts(ps.ng.tax_most_pre_post_stroke, function(x) x / sum(x))#plot_bar(ps.ng.tax_most_SampleType_, fill = "Phylum") + geom_bar(aes(color=Phylum, fill=Phylum), stat="identity", position="stack")plot_bar(ps.ng.tax_most_pre_post_stroke_, fill="Phylum") + geom_bar(aes(), stat="identity", position="stack") +scale_fill_manual(values = c("darkblue", "darkgoldenrod1", "darkseagreen", "darkorchid", "darkolivegreen1", "lightskyblue", "darkgreen", "deeppink", "khaki2", "firebrick", "brown1", "darkorange1", "cyan1", "royalblue4", "darksalmon", "darkblue","royalblue4", "dodgerblue3", "steelblue1", "lightskyblue", "darkseagreen", "darkgoldenrod1", "darkseagreen", "darkorchid", "darkolivegreen1", "brown1", "darkorange1", "cyan1", "darkgrey")) + theme(axis.text = element_text(size = theme.size, colour="black"))```\pagebreakUse color according to phylum. Do separate panels Stroke and Sex_age.```{r, echo=TRUE, warning=FALSE}ps.ng.tax_most_copied <- data.table::copy(ps.ng.tax_most_)#FITTING6: regulate the bar height if it has replicates: 5+6+6+8+4+8+6+7=25+25=50otu_table(ps.ng.tax_most_)[,c("1")] <- otu_table(ps.ng.tax_most_)[,c("1")]/5otu_table(ps.ng.tax_most_)[,c("2")] <- otu_table(ps.ng.tax_most_)[,c("2")]/5otu_table(ps.ng.tax_most_)[,c("5")] <- otu_table(ps.ng.tax_most_)[,c("5")]/5otu_table(ps.ng.tax_most_)[,c("6")] <- otu_table(ps.ng.tax_most_)[,c("6")]/5otu_table(ps.ng.tax_most_)[,c("7")] <- otu_table(ps.ng.tax_most_)[,c("7")]/5otu_table(ps.ng.tax_most_)[,c("8")] <- otu_table(ps.ng.tax_most_)[,c("8")]/6otu_table(ps.ng.tax_most_)[,c("9")] <- otu_table(ps.ng.tax_most_)[,c("9")]/6otu_table(ps.ng.tax_most_)[,c("10")] <- otu_table(ps.ng.tax_most_)[,c("10")]/6otu_table(ps.ng.tax_most_)[,c("12")] <- otu_table(ps.ng.tax_most_)[,c("12")]/6otu_table(ps.ng.tax_most_)[,c("13")] <- otu_table(ps.ng.tax_most_)[,c("13")]/6otu_table(ps.ng.tax_most_)[,c("14")] <- otu_table(ps.ng.tax_most_)[,c("14")]/6otu_table(ps.ng.tax_most_)[,c("15")] <- otu_table(ps.ng.tax_most_)[,c("15")]/6otu_table(ps.ng.tax_most_)[,c("16")] <- otu_table(ps.ng.tax_most_)[,c("16")]/6otu_table(ps.ng.tax_most_)[,c("17")] <- otu_table(ps.ng.tax_most_)[,c("17")]/6otu_table(ps.ng.tax_most_)[,c("18")] <- otu_table(ps.ng.tax_most_)[,c("18")]/6otu_table(ps.ng.tax_most_)[,c("19")] <- otu_table(ps.ng.tax_most_)[,c("19")]/6otu_table(ps.ng.tax_most_)[,c("20")] <- otu_table(ps.ng.tax_most_)[,c("20")]/6otu_table(ps.ng.tax_most_)[,c("21")] <- otu_table(ps.ng.tax_most_)[,c("21")]/8otu_table(ps.ng.tax_most_)[,c("22")] <- otu_table(ps.ng.tax_most_)[,c("22")]/8otu_table(ps.ng.tax_most_)[,c("23")] <- otu_table(ps.ng.tax_most_)[,c("23")]/8otu_table(ps.ng.tax_most_)[,c("24")] <- otu_table(ps.ng.tax_most_)[,c("24")]/8otu_table(ps.ng.tax_most_)[,c("25")] <- otu_table(ps.ng.tax_most_)[,c("25")]/8otu_table(ps.ng.tax_most_)[,c("26")] <- otu_table(ps.ng.tax_most_)[,c("26")]/8otu_table(ps.ng.tax_most_)[,c("27")] <- otu_table(ps.ng.tax_most_)[,c("27")]/8otu_table(ps.ng.tax_most_)[,c("28")] <- otu_table(ps.ng.tax_most_)[,c("28")]/8otu_table(ps.ng.tax_most_)[,c("29")] <- otu_table(ps.ng.tax_most_)[,c("29")]/4otu_table(ps.ng.tax_most_)[,c("30")] <- otu_table(ps.ng.tax_most_)[,c("30")]/4otu_table(ps.ng.tax_most_)[,c("31")] <- otu_table(ps.ng.tax_most_)[,c("31")]/4otu_table(ps.ng.tax_most_)[,c("32")] <- otu_table(ps.ng.tax_most_)[,c("32")]/4otu_table(ps.ng.tax_most_)[,c("33")] <- otu_table(ps.ng.tax_most_)[,c("33")]/8otu_table(ps.ng.tax_most_)[,c("34")] <- otu_table(ps.ng.tax_most_)[,c("34")]/8otu_table(ps.ng.tax_most_)[,c("35")] <- otu_table(ps.ng.tax_most_)[,c("35")]/8otu_table(ps.ng.tax_most_)[,c("36")] <- otu_table(ps.ng.tax_most_)[,c("36")]/8otu_table(ps.ng.tax_most_)[,c("37")] <- otu_table(ps.ng.tax_most_)[,c("37")]/8otu_table(ps.ng.tax_most_)[,c("38")] <- otu_table(ps.ng.tax_most_)[,c("38")]/8otu_table(ps.ng.tax_most_)[,c("39")] <- otu_table(ps.ng.tax_most_)[,c("39")]/8otu_table(ps.ng.tax_most_)[,c("51")] <- otu_table(ps.ng.tax_most_)[,c("51")]/8otu_table(ps.ng.tax_most_)[,c("40")] <- otu_table(ps.ng.tax_most_)[,c("40")]/6otu_table(ps.ng.tax_most_)[,c("41")] <- otu_table(ps.ng.tax_most_)[,c("41")]/6otu_table(ps.ng.tax_most_)[,c("42")] <- otu_table(ps.ng.tax_most_)[,c("42")]/6otu_table(ps.ng.tax_most_)[,c("43")] <- otu_table(ps.ng.tax_most_)[,c("43")]/6otu_table(ps.ng.tax_most_)[,c("44")] <- otu_table(ps.ng.tax_most_)[,c("44")]/6otu_table(ps.ng.tax_most_)[,c("46")] <- otu_table(ps.ng.tax_most_)[,c("46")]/6otu_table(ps.ng.tax_most_)[,c("47")] <- otu_table(ps.ng.tax_most_)[,c("47")]/7otu_table(ps.ng.tax_most_)[,c("48")] <- otu_table(ps.ng.tax_most_)[,c("48")]/7otu_table(ps.ng.tax_most_)[,c("49")] <- otu_table(ps.ng.tax_most_)[,c("49")]/7otu_table(ps.ng.tax_most_)[,c("50")] <- otu_table(ps.ng.tax_most_)[,c("50")]/7otu_table(ps.ng.tax_most_)[,c("52")] <- otu_table(ps.ng.tax_most_)[,c("52")]/7otu_table(ps.ng.tax_most_)[,c("53")] <- otu_table(ps.ng.tax_most_)[,c("53")]/7otu_table(ps.ng.tax_most_)[,c("55")] <- otu_table(ps.ng.tax_most_)[,c("55")]/7#plot_bar(ps.ng.tax_most_swab_, x="Phylum", fill = "Phylum", facet_grid = Patient~RoundDay) + geom_bar(aes(color=Phylum, fill=Phylum), stat="identity", position="stack") + theme(axis.text = element_text(size = theme.size, colour="black"))plot_bar(ps.ng.tax_most_, x="Phylum", fill="Phylum", facet_grid = pre_post_stroke~Sex_age) + geom_bar(aes(), stat="identity", position="stack") +scale_fill_manual(values = c("darkblue", "darkgoldenrod1", "darkseagreen", "darkorchid", "darkolivegreen1", "lightskyblue", "darkgreen", "deeppink", "khaki2", "firebrick", "brown1", "darkorange1", "cyan1", "royalblue4", "darksalmon", "darkblue","royalblue4", "dodgerblue3", "steelblue1", "lightskyblue", "darkseagreen", "darkgoldenrod1", "darkseagreen", "darkorchid", "darkolivegreen1", "brown1", "darkorange1", "cyan1", "darkgrey")) + theme(axis.text = element_text(size = 5, colour="black"), axis.text.x=element_blank(), axis.ticks=element_blank()) + theme(legend.position="bottom") + guides(fill=guide_legend(nrow=2))``````{r, echo=FALSE, warning=FALSE}#knitr::include_graphics("./Phyloseq_files/figure-html/unnamed-chunk-10-1.png")#> tax_table(carbom)#Taxonomy Table: [205 taxa by 7 taxonomic ranks]:# Domain Supergroup Division Class#Otu001 "Eukaryota" "Archaeplastida" "Chlorophyta" "Mamiellophyceae"# Order Family Genus#Otu001 "Mamiellales" "Bathycoccaceae" "Ostreococcus"#sample_data(ps.ng.tax)```## Bar plots in class level```{r, echo=TRUE, warning=FALSE}plot_bar(ps.ng.tax_most_copied, fill="Class") + geom_bar(aes(), stat="identity", position="stack") +scale_fill_manual(values = c("darkblue", "darkgoldenrod1", "darkseagreen", "darkorchid", "darkolivegreen1", "lightskyblue", "darkgreen", "deeppink", "khaki2", "firebrick", "brown1", "darkorange1", "cyan1", "royalblue4", "darksalmon", "darkblue","royalblue4", "dodgerblue3", "steelblue1", "lightskyblue", "darkseagreen", "darkgoldenrod1", "darkseagreen", "darkorchid", "darkolivegreen1", "brown1", "darkorange1", "cyan1", "darkgrey")) + theme(axis.text = element_text(size = 5, colour="black")) + theme(legend.position="bottom") + guides(fill=guide_legend(nrow=3))```Regroup together pre vs post stroke samples and normalize number of reads in each group using median sequencing depth.```{r, echo=TRUE, warning=FALSE}plot_bar(ps.ng.tax_most_pre_post_stroke_, fill="Class") + geom_bar(aes(), stat="identity", position="stack") +scale_fill_manual(values = c("darkblue", "darkgoldenrod1", "darkseagreen", "darkorchid", "darkolivegreen1", "lightskyblue", "darkgreen", "deeppink", "khaki2", "firebrick", "brown1", "darkorange1", "cyan1", "royalblue4", "darksalmon", "darkblue","royalblue4", "dodgerblue3", "steelblue1", "lightskyblue", "darkseagreen", "darkgoldenrod1", "darkseagreen", "darkorchid", "darkolivegreen1", "brown1", "darkorange1", "cyan1", "darkgrey")) + theme(axis.text = element_text(size = theme.size, colour="black"))```\pagebreakUse color according to class. Do separate panels Stroke and Sex_age.```{r, echo=TRUE, warning=FALSE}#-- If existing replicates, to be processed as follows --plot_bar(ps.ng.tax_most_, x="Class", fill="Class", facet_grid = pre_post_stroke~Sex_age) + geom_bar(aes(), stat="identity", position="stack") +scale_fill_manual(values = c("darkblue", "darkgoldenrod1", "darkseagreen", "darkorchid", "darkolivegreen1", "lightskyblue", "darkgreen", "deeppink", "khaki2", "firebrick", "brown1", "darkorange1", "cyan1", "royalblue4", "darksalmon", "darkblue","royalblue4", "dodgerblue3", "steelblue1", "lightskyblue", "darkseagreen", "darkgoldenrod1", "darkseagreen", "darkorchid", "darkolivegreen1", "brown1", "darkorange1", "cyan1", "darkgrey")) + theme(axis.text = element_text(size = 5, colour="black"), axis.text.x=element_blank(), axis.ticks=element_blank()) + theme(legend.position="bottom") + guides(fill=guide_legend(nrow=3))```## Bar plots in order level```{r, echo=TRUE, warning=FALSE}plot_bar(ps.ng.tax_most_copied, fill="Order") + geom_bar(aes(), stat="identity", position="stack") +scale_fill_manual(values = c("darkblue", "darkgoldenrod1", "darkseagreen", "darkorchid", "darkolivegreen1", "lightskyblue", "darkgreen", "deeppink", "khaki2", "firebrick", "brown1", "darkorange1", "cyan1", "royalblue4", "darksalmon", "darkblue","royalblue4", "dodgerblue3", "steelblue1", "lightskyblue", "darkseagreen", "darkgoldenrod1", "darkseagreen", "darkorchid", "darkolivegreen1", "brown1", "darkorange1", "cyan1", "darkgrey")) + theme(axis.text = element_text(size = 5, colour="black")) + theme(legend.position="bottom") + guides(fill=guide_legend(nrow=4))```Regroup together pre vs post stroke and normalize number of reads in each group using median sequencing depth.```{r, echo=TRUE, warning=FALSE}plot_bar(ps.ng.tax_most_pre_post_stroke_, fill="Order") + geom_bar(aes(), stat="identity", position="stack") +scale_fill_manual(values = c("darkblue", "darkgoldenrod1", "darkseagreen", "darkorchid", "darkolivegreen1", "lightskyblue", "darkgreen", "deeppink", "khaki2", "firebrick", "brown1", "darkorange1", "cyan1", "royalblue4", "darksalmon", "darkblue","royalblue4", "dodgerblue3", "steelblue1", "lightskyblue", "darkseagreen", "darkgoldenrod1", "darkseagreen", "darkorchid", "darkolivegreen1", "brown1", "darkorange1", "cyan1", "darkgrey")) + theme(axis.text = element_text(size = theme.size, colour="black")) + theme(legend.position="bottom") + guides(fill=guide_legend(nrow=4))```\pagebreakUse color according to order. Do separate panels Stroke and Sex_age.```{r, echo=TRUE, warning=FALSE}#FITTING7: regulate the bar height if it has replicatesplot_bar(ps.ng.tax_most_, x="Order", fill="Order", facet_grid = pre_post_stroke~Sex_age) + geom_bar(aes(), stat="identity", position="stack") +scale_fill_manual(values = c("darkblue", "darkgoldenrod1", "darkseagreen", "darkorchid", "darkolivegreen1", "lightskyblue", "darkgreen", "deeppink", "khaki2", "firebrick", "brown1", "darkorange1", "cyan1", "royalblue4", "darksalmon", "darkblue","royalblue4", "dodgerblue3", "steelblue1", "lightskyblue", "darkseagreen", "darkgoldenrod1", "darkseagreen", "darkorchid", "darkolivegreen1", "brown1", "darkorange1", "cyan1", "darkgrey")) + theme(axis.text = element_text(size = 5, colour="black"), axis.text.x=element_blank(), axis.ticks=element_blank()) + theme(legend.position="bottom") + guides(fill=guide_legend(nrow=4))```## Bar plots in family level```{r, echo=TRUE, warning=FALSE}plot_bar(ps.ng.tax_most_copied, fill="Family") + geom_bar(aes(), stat="identity", position="stack") +scale_fill_manual(values = c("#FF0000", "#000000", "#0000FF", "#C0C0C0", "#FFFFFF", "#FFFF00", "#00FFFF", "#FFA500", "#00FF00", "#808080", "#FF00FF", "#800080", "#FDD017", "#0000A0", "#3BB9FF", "#008000", "#800000", "#ADD8E6", "#F778A1", "#800517", "#736F6E", "#F52887", "#C11B17", "#5CB3FF", "#A52A2A", "#FF8040", "#2B60DE", "#736AFF", "#1589FF", "#98AFC7", "#8D38C9", "#307D7E", "#F6358A", "#151B54", "#6D7B8D", "#FDEEF4", "#FF0080", "#F88017", "#2554C7", "#FFF8C6", "#D4A017", "#306EFF", "#151B8D", "#9E7BFF", "#EAC117", "#E0FFFF", "#15317E", "#6C2DC7", "#FBB917", "#FCDFFF", "#15317E", "#254117", "#FAAFBE", "#357EC7")) + theme(axis.text = element_text(size = 5, colour="black")) + theme(legend.position="bottom") + guides(fill=guide_legend(nrow=8))```Regroup together pre vs post stroke samples and normalize number of reads in each group using median sequencing depth.```{r, echo=TRUE, warning=FALSE}plot_bar(ps.ng.tax_most_pre_post_stroke_, fill="Family") + geom_bar(aes(), stat="identity", position="stack") +scale_fill_manual(values = c("#FF0000", "#000000", "#0000FF", "#C0C0C0", "#FFFFFF", "#FFFF00", "#00FFFF", "#FFA500", "#00FF00", "#808080", "#FF00FF", "#800080", "#FDD017", "#0000A0", "#3BB9FF", "#008000", "#800000", "#ADD8E6", "#F778A1", "#800517", "#736F6E", "#F52887", "#C11B17", "#5CB3FF", "#A52A2A", "#FF8040", "#2B60DE", "#736AFF", "#1589FF", "#98AFC7", "#8D38C9", "#307D7E", "#F6358A", "#151B54", "#6D7B8D", "#FDEEF4", "#FF0080", "#F88017", "#2554C7", "#FFF8C6", "#D4A017", "#306EFF", "#151B8D", "#9E7BFF", "#EAC117", "#E0FFFF", "#15317E", "#6C2DC7", "#FBB917", "#FCDFFF", "#15317E", "#254117", "#FAAFBE", "#357EC7")) + theme(axis.text = element_text(size = theme.size, colour="black")) + theme(legend.position="bottom") + guides(fill=guide_legend(nrow=8))```\pagebreakUse color according to family. Do separate panels Stroke and Sex_age.```{r, echo=TRUE, warning=FALSE}#-- If existing replicates, to be processed as follows --plot_bar(ps.ng.tax_most_, x="Family", fill="Family", facet_grid = pre_post_stroke~Sex_age) + geom_bar(aes(), stat="identity", position="stack") +scale_fill_manual(values = c("#FF0000", "#000000", "#0000FF", "#C0C0C0", "#FFFFFF", "#FFFF00", "#00FFFF", "#FFA500", "#00FF00", "#808080", "#FF00FF", "#800080", "#FDD017", "#0000A0", "#3BB9FF", "#008000", "#800000", "#ADD8E6", "#F778A1", "#800517", "#736F6E", "#F52887", "#C11B17", "#5CB3FF", "#A52A2A", "#FF8040", "#2B60DE", "#736AFF", "#1589FF", "#98AFC7", "#8D38C9", "#307D7E", "#F6358A", "#151B54", "#6D7B8D", "#FDEEF4", "#FF0080", "#F88017", "#2554C7", "#FFF8C6", "#D4A017", "#306EFF", "#151B8D", "#9E7BFF", "#EAC117", "#E0FFFF", "#15317E", "#6C2DC7", "#FBB917", "#FCDFFF", "#15317E", "#254117", "#FAAFBE", "#357EC7")) + theme(axis.text = element_text(size = 5, colour="black"), axis.text.x=element_blank(), axis.ticks=element_blank()) + theme(legend.position="bottom") + guides(fill=guide_legend(nrow=8))``````{r, echo=FALSE, warning=FALSE}#MOVE_FROM_ABOVE: ## Bar plots in genus level#MOVE_FROM_ABOVE: Regroup together pre vs post stroke samples and normalize number of reads in each group using median sequencing depth.#plot_bar(ps.ng.tax_most_pre_post_stroke_, fill="Genus") + geom_bar(aes(), stat="identity", position="stack") +#scale_fill_manual(values = c("#FF0000", "#000000", "#0000FF", "#C0C0C0", "#FFFFFF", "#FFFF00", "#00FFFF", "#FFA500", "#00FF00", "#808080", "#FF00FF", "#800080", "#FDD017", "#0000A0", "#3BB9FF", "#008000", "#800000", "#ADD8E6", "#F778A1", "#800517", "#736F6E", "#F52887", "#C11B17", "#5CB3FF", "#A52A2A", "#FF8040", "#2B60DE", "#736AFF", "#1589FF", "#98AFC7", "#8D38C9", "#307D7E", "#F6358A", "#151B54", "#6D7B8D", "#FDEEF4", "#FF0080", "#F88017", "#2554C7", "#FFF8C6", "#D4A017", "#306EFF", "#151B8D", "#9E7BFF", "#EAC117", "#E0FFFF", "#15317E", "#6C2DC7", "#FBB917", "#FCDFFF", "#15317E", "#254117", "#FAAFBE", "#357EC7")) + theme(axis.text = element_text(size = theme.size, colour="black")) + theme(legend.position="bottom")```\pagebreak```{r, echo=FALSE, warning=FALSE}#MOVE_FROM_ABOVE: Use color according to genus. Do separate panels Stroke and Sex_age.##-- If existing replicates, to be processed as follows --#plot_bar(ps.ng.tax_most_, x="Genus", fill="Genus", facet_grid = pre_post_stroke~Sex_age) + geom_bar(aes(), stat="identity", position="stack") +#scale_fill_manual(values = c("#FF0000", "#000000", "#0000FF", "#C0C0C0", "#FFFFFF", "#FFFF00", "#00FFFF", "#FFA500", "#00FF00", "#808080", "#FF00FF", "#800080", "#FDD017", "#0000A0", "#3BB9FF", "#008000", "#800000", "#ADD8E6", "#F778A1", "#800517", "#736F6E", "#F52887", "#C11B17", "#5CB3FF", "#A52A2A", "#FF8040", "#2B60DE", "#736AFF", "#1589FF", "#98AFC7", "#8D38C9", "#307D7E", "#F6358A", "#151B54", "#6D7B8D", "#FDEEF4", "#FF0080", "#F88017", "#2554C7", "#FFF8C6", "#D4A017", "#306EFF", "#151B8D", "#9E7BFF", "#EAC117", "#E0FFFF", "#15317E", "#6C2DC7", "#FBB917", "#FCDFFF", "#15317E", "#254117", "#FAAFBE", "#357EC7")) + theme(axis.text = element_text(size = 6, colour="black"), axis.text.x=element_blank(), axis.ticks=element_blank()) + theme(legend.position="bottom") + guides(fill=guide_legend(nrow=18))```\pagebreak# Alpha diversityPlot Chao1 richness estimator, Observed OTUs, Shannon index, and Phylogenetic diversity.Regroup together samples from the same group.```{r, echo=FALSE, warning=FALSE}# using rarefied data#FITTING2: CONSOLE:#gunzip table_even4753.biom.gz#alpha_diversity.py -i table_even42369.biom --metrics chao1,observed_otus,shannon,PD_whole_tree -o adiv_even.txt -t ../clustering/rep_set.tre#gunzip table_even4753.biom.gz#alpha_diversity.py -i table_even4753.biom --metrics chao1,observed_otus,shannon,PD_whole_tree -o adiv_even.txt -t ../clustering_stool/rep_set.tre#gunzip table_even4753.biom.gz#alpha_diversity.py -i table_even4753.biom --metrics chao1,observed_otus,shannon,PD_whole_tree -o adiv_even.txt -t ../clustering_swab/rep_set.tre``````{r, echo=TRUE, warning=FALSE}hmp.div_qiime <- read.csv("adiv_even.txt", sep="\t")colnames(hmp.div_qiime) <- c("sam_name", "chao1", "observed_otus", "shannon", "PD_whole_tree")row.names(hmp.div_qiime) <- hmp.div_qiime$sam_namediv.df <- merge(hmp.div_qiime, hmp.meta, by = "sam_name")div.df2 <- div.df[, c("Group", "chao1", "shannon", "observed_otus", "PD_whole_tree")]colnames(div.df2) <- c("Group", "Chao-1", "Shannon", "OTU", "Phylogenetic Diversity")#colnames(div.df2)options(max.print=999999)#27 H47 830.5000 5.008482 319 10.60177#FITTING4: if occuring "Computation failed in `stat_signif()`:not enough 'y' observations"#means: the patient H47 contains only one sample, it should be removed for the statistical p-values calculations.#delete H47(1)#div.df2 <- div.df2[-c(3), ]#div.df2 <- div.df2[-c(55,54, 45,40,39,27,26,25,1), ]knitr::kable(div.df2) %>% kable_styling(bootstrap_options = c("striped", "hover", "condensed", "responsive"))#https://uc-r.github.io/t_test#We can perform the test with t.test and transform our data and we can also perform the nonparametric test with the wilcox.test function.stat.test.Shannon <- compare_means(Shannon ~ Group, data = div.df2,method = "t.test")knitr::kable(stat.test.Shannon) %>% kable_styling(bootstrap_options = c("striped", "hover", "condensed", "responsive"))div_df_melt <- reshape2::melt(div.df2)#head(div_df_melt)#https://plot.ly/r/box-plots/#horizontal-boxplot#http://www.sthda.com/english/wiki/print.php?id=177#https://rpkgs.datanovia.com/ggpubr/reference/as_ggplot.html#http://www.sthda.com/english/articles/24-ggpubr-publication-ready-plots/82-ggplot2-easy-way-to-change-graphical-parameters/#https://plot.ly/r/box-plots/#horizontal-boxplot#library("gridExtra")#par(mfrow=c(4,1))p <- ggboxplot(div_df_melt, x = "Group", y = "value",facet.by = "variable",scales = "free",width = 0.5,fill = "gray", legend= "right")#ggpar(p, xlab = FALSE, ylab = FALSE)lev <- levels(factor(div_df_melt$Group)) # get the variables#FITTING4: delete H47(1) in lev#lev <- lev[-c(3)]# make a pairwise list that we want to compare.#my_stat_compare_means#https://stackoverflow.com/questions/47839988/indicating-significance-with-ggplot2-in-a-boxplot-with-multiple-groupsL.pairs <- combn(seq_along(lev), 2, simplify = FALSE, FUN = function(i) lev[i]) #%>% filter(p.signif != "ns")my_stat_compare_means <- function (mapping = NULL, data = NULL, method = NULL, paired = FALSE,method.args = list(), ref.group = NULL, comparisons = NULL,hide.ns = FALSE, label.sep = ", ", label = NULL, label.x.npc = "left",label.y.npc = "top", label.x = NULL, label.y = NULL, tip.length = 0.03,symnum.args = list(), geom = "text", position = "identity",na.rm = FALSE, show.legend = NA, inherit.aes = TRUE, ...){if (!is.null(comparisons)) {method.info <- ggpubr:::.method_info(method)method <- method.info$methodmethod.args <- ggpubr:::.add_item(method.args, paired = paired)if (method == "wilcox.test")method.args$exact <- FALSEpms <- list(...)size <- ifelse(is.null(pms$size), 0.3, pms$size)color <- ifelse(is.null(pms$color), "black", pms$color)map_signif_level <- FALSEif (is.null(label))label <- "p.format"if (ggpubr:::.is_p.signif_in_mapping(mapping) | (label %in% "p.signif")) {if (ggpubr:::.is_empty(symnum.args)) {map_signif_level <- c(`****` = 1e-04, `***` = 0.001,`**` = 0.01, `*` = 0.05, ns = 1)} else {map_signif_level <- symnum.args}if (hide.ns)names(map_signif_level)[5] <- " "}step_increase <- ifelse(is.null(label.y), 0.12, 0)ggsignif::geom_signif(comparisons = comparisons, y_position = label.y,test = method, test.args = method.args, step_increase = step_increase,size = size, color = color, map_signif_level = map_signif_level,tip_length = tip.length, data = data)} else {mapping <- ggpubr:::.update_mapping(mapping, label)layer(stat = StatCompareMeans, data = data, mapping = mapping,geom = geom, position = position, show.legend = show.legend,inherit.aes = inherit.aes, params = list(label.x.npc = label.x.npc,label.y.npc = label.y.npc, label.x = label.x,label.y = label.y, label.sep = label.sep, method = method,method.args = method.args, paired = paired, ref.group = ref.group,symnum.args = symnum.args, hide.ns = hide.ns,na.rm = na.rm, ...))}}p2 <- p +stat_compare_means(method="t.test",comparisons = list(c("Group1", "Group2"), c("Group1", "Group3"), c("Group1", "Group4"), c("Group1", "Group6"), c("Group1", "Group8"), c("Group2", "Group5"),c("Group4", "Group5"),c("Group4", "Group6"),c("Group4", "Group7"),c("Group6", "Group7")),label = "p.signif",symnum.args <- list(cutpoints = c(0, 0.0001, 0.001, 0.01, 0.05, 1), symbols = c("****", "***", "**", "*", "ns")))#comparisons = L.pairs,#symnum.args <- list(cutpoints = c(0, 0.0001, 0.001, 0.01, 0.05), symbols = c("****", "***", "**", "*")),#stat_pvalue_manual#print(p2)#https://stackoverflow.com/questions/20500706/saving-multiple-ggplots-from-ls-into-one-and-separate-files-in-r#FITTING3: mkdir figuresggsave("./figures/alpha_diversity_Group.png", device="png", height = 10, width = 12)ggsave("./figures/alpha_diversity_Group.svg", device="svg", height = 10, width = 12)p3 <- p +stat_compare_means(method="t.test",comparisons = list(c("Group2", "Group4"), c("Group2", "Group6"), c("Group4", "Group8"), c("Group6", "Group8")),label = "p.signif",symnum.args <- list(cutpoints = c(0, 0.0001, 0.001, 0.01, 0.05, 1), symbols = c("****", "***", "**", "*", "ns")),)#symnum.args <- list(cutpoints = c(0, 0.0001, 0.001, 0.01, 0.05), symbols = c("****", "***", "**", "*")),#stat_pvalue_manual#print(p2)#https://stackoverflow.com/questions/20500706/saving-multiple-ggplots-from-ls-into-one-and-separate-files-in-r#FITTING3: mkdir figuresggsave("./figures/alpha_diversity_Group2.png", device="png", height = 10, width = 12)ggsave("./figures/alpha_diversity_Group2.svg", device="svg", height = 10, width = 12)```# Selected alpha diversity```{r, echo=TRUE, warning=FALSE, fig.cap="Alpha diversity", out.width = '100%', fig.align= "center"}knitr::include_graphics("./figures/alpha_diversity_Group2.png")selected_alpha_diversities<-read.csv("selected_alpha_diversities.txt",sep="\t")knitr::kable(selected_alpha_diversities) %>% kable_styling(bootstrap_options = c("striped", "hover", "condensed", "responsive"))```# Beta diversity```{r, echo=TRUE, warning=FALSE, fig.cap="Beta diversity", out.width = '100%', fig.align= "center"}#file:///home/jhuang/DATA/Data_Marius_16S/core_diversity_e42369/bdiv_even42369_Group/unweighted_unifrac_boxplots/Group_Stats.txtbeta_diversity_group_stats<-read.csv("unweighted_unifrac_boxplots_Group_Stats.txt",sep="\t")knitr::kable(beta_diversity_group_stats) %>% kable_styling(bootstrap_options = c("striped", "hover", "condensed", "responsive"))#NOTE: Run this Phyloseq0.Rmd, then run the code of MicrobiotaProcess.R to manually generate PCoA.png, then run this Phyloseq.Rmd!#NOTE: AT_FIRST_DEACTIVATE_THIS_LINE: knitr::include_graphics("./figures/PCoA.png")```# Differential abundance analysisDifferential abundance analysis aims to find the differences in the abundance of each taxa between two groups of samples, assigning a significance value to each comparison.## Group2 vs Group4```{r, echo=TRUE, warning=FALSE}library("DESeq2")#ALTERNATIVE using ps.ng.tax_most_copied: ps.ng.tax (40594) vs. ps.ng.tax_most_copied (166)ps.ng.tax_sel <- ps.ng.tax#FITTING5: correct the id of the group members, see FITTING6otu_table(ps.ng.tax_sel) <- otu_table(ps.ng.tax)[,c("8","9","10","12","13","14", "21","22","23","24","25","26","27","28")]diagdds = phyloseq_to_deseq2(ps.ng.tax_sel, ~Group)diagdds$Group <- relevel(diagdds$Group, "Group4")diagdds = DESeq(diagdds, test="Wald", fitType="parametric")resultsNames(diagdds)res = results(diagdds, cooksCutoff = FALSE)alpha = 0.05sigtab = res[which(res$padj < alpha), ]sigtab = cbind(as(sigtab, "data.frame"), as(tax_table(ps.ng.tax_sel)[rownames(sigtab), ], "matrix"))sigtab <- sigtab[rownames(sigtab) %in% rownames(tax_table(ps.ng.tax_most_copied)), ]kable(sigtab) %>%kable_styling(bootstrap_options = c("striped", "hover", "condensed", "responsive"))#rownames(sigtab) %in% rownames(tax_table(ps.ng.tax_most_copied))#subv %in% v### returns a vector TRUE FALSE#is.element(subv, v)### returns a vector TRUE FALSElibrary("ggplot2")theme_set(theme_bw())scale_fill_discrete <- function(palname = "Set1", ...) {scale_fill_brewer(palette = palname, ...)}x = tapply(sigtab$log2FoldChange, sigtab$Order, function(x) max(x))x = sort(x)sigtab$Order = factor(as.character(sigtab$Order), levels=names(x))x = tapply(sigtab$log2FoldChange, sigtab$Family, function(x) max(x))x = sort(x)sigtab$Family = factor(as.character(sigtab$Family), levels=names(x))ggplot(sigtab, aes(x=log2FoldChange, y=Family, color=Order)) + geom_point(aes(size=padj)) + scale_size_continuous(name="padj",range=c(8,4))+theme(axis.text.x = element_text(angle = -25, hjust = 0, vjust=0.5))#Error in checkForExperimentalReplicates(object, modelMatrix) :# The design matrix has the same number of samples and coefficients to fit,# so estimation of dispersion is not possible. Treating samples# as replicates was deprecated in v1.20 and no longer supported since v1.22.```## Group2 vs Group6```{r, echo=TRUE, warning=FALSE}ps.ng.tax_sel <- ps.ng.taxotu_table(ps.ng.tax_sel) <- otu_table(ps.ng.tax)[,c("8","9","10","12","13","14", "33","34","35","36","37","38","39","51")]diagdds = phyloseq_to_deseq2(ps.ng.tax_sel, ~Group)diagdds$Group <- relevel(diagdds$Group, "Group6")diagdds = DESeq(diagdds, test="Wald", fitType="parametric")resultsNames(diagdds)res = results(diagdds, cooksCutoff = FALSE)alpha = 0.05sigtab = res[which(res$padj < alpha), ]sigtab = cbind(as(sigtab, "data.frame"), as(tax_table(ps.ng.tax_sel)[rownames(sigtab), ], "matrix"))sigtab <- sigtab[rownames(sigtab) %in% rownames(tax_table(ps.ng.tax_most_copied)), ]kable(sigtab) %>%kable_styling(bootstrap_options = c("striped", "hover", "condensed", "responsive"))library("ggplot2")theme_set(theme_bw())scale_fill_discrete <- function(palname = "Set1", ...) {scale_fill_brewer(palette = palname, ...)}x = tapply(sigtab$log2FoldChange, sigtab$Order, function(x) max(x))x = sort(x)sigtab$Order = factor(as.character(sigtab$Order), levels=names(x))x = tapply(sigtab$log2FoldChange, sigtab$Family, function(x) max(x))x = sort(x)sigtab$Family = factor(as.character(sigtab$Family), levels=names(x))ggplot(sigtab, aes(x=log2FoldChange, y=Family, color=Order)) + geom_point(aes(size=padj)) + scale_size_continuous(name="padj",range=c(8,4))+theme(axis.text.x = element_text(angle = -25, hjust = 0, vjust=0.5))```## Group4 vs Group8```{r, echo=TRUE, warning=FALSE}ps.ng.tax_sel <- ps.ng.taxotu_table(ps.ng.tax_sel) <- otu_table(ps.ng.tax)[,c("21","22","23","24","25","26","27","28", "47","48","49","50","52","53","55")]diagdds = phyloseq_to_deseq2(ps.ng.tax_sel, ~Group)diagdds$Group <- relevel(diagdds$Group, "Group8")diagdds = DESeq(diagdds, test="Wald", fitType="parametric")resultsNames(diagdds)res = results(diagdds, cooksCutoff = FALSE)alpha = 0.05sigtab = res[which(res$padj < alpha), ]sigtab = cbind(as(sigtab, "data.frame"), as(tax_table(ps.ng.tax_sel)[rownames(sigtab), ], "matrix"))sigtab <- sigtab[rownames(sigtab) %in% rownames(tax_table(ps.ng.tax_most_copied)), ]kable(sigtab) %>%kable_styling(bootstrap_options = c("striped", "hover", "condensed", "responsive"))library("ggplot2")theme_set(theme_bw())scale_fill_discrete <- function(palname = "Set1", ...) {scale_fill_brewer(palette = palname, ...)}x = tapply(sigtab$log2FoldChange, sigtab$Order, function(x) max(x))x = sort(x)sigtab$Order = factor(as.character(sigtab$Order), levels=names(x))x = tapply(sigtab$log2FoldChange, sigtab$Family, function(x) max(x))x = sort(x)sigtab$Family = factor(as.character(sigtab$Family), levels=names(x))ggplot(sigtab, aes(x=log2FoldChange, y=Family, color=Order)) + geom_point(aes(size=padj)) + scale_size_continuous(name="padj",range=c(8,4))+theme(axis.text.x = element_text(angle = -25, hjust = 0, vjust=0.5))```## Group6 vs Group8```{r, echo=TRUE, warning=FALSE}ps.ng.tax_sel <- ps.ng.taxotu_table(ps.ng.tax_sel) <- otu_table(ps.ng.tax)[,c("33","34","35","36","37","38","39","51", "47","48","49","50","52","53","55")]diagdds = phyloseq_to_deseq2(ps.ng.tax_sel, ~Group)diagdds$Group <- relevel(diagdds$Group, "Group8")diagdds = DESeq(diagdds, test="Wald", fitType="parametric")resultsNames(diagdds)res = results(diagdds, cooksCutoff = FALSE)alpha = 0.05sigtab = res[which(res$padj < alpha), ]sigtab = cbind(as(sigtab, "data.frame"), as(tax_table(ps.ng.tax_sel)[rownames(sigtab), ], "matrix"))sigtab <- sigtab[rownames(sigtab) %in% rownames(tax_table(ps.ng.tax_most_copied)), ]kable(sigtab) %>%kable_styling(bootstrap_options = c("striped", "hover", "condensed", "responsive"))library("ggplot2")theme_set(theme_bw())scale_fill_discrete <- function(palname = "Set1", ...) {scale_fill_brewer(palette = palname, ...)}x = tapply(sigtab$log2FoldChange, sigtab$Order, function(x) max(x))x = sort(x)sigtab$Order = factor(as.character(sigtab$Order), levels=names(x))x = tapply(sigtab$log2FoldChange, sigtab$Family, function(x) max(x))x = sort(x)sigtab$Family = factor(as.character(sigtab$Family), levels=names(x))ggplot(sigtab, aes(x=log2FoldChange, y=Family, color=Order)) + geom_point(aes(size=padj)) + scale_size_continuous(name="padj",range=c(8,4))+theme(axis.text.x = element_text(angle = -25, hjust = 0, vjust=0.5))```
点赞本文的读者
还没有人对此文章表态
本文有评论
没有评论
看文章,发评论,不要沉默
最受欢迎文章
- Motif Discovery in Biological Sequences: A Comparison of MEME and HOMER
- Why Do Significant Gene Lists Change After Adding Additional Conditions in Differential Gene Expression Analysis?
- Calling peaks using findPeaks of HOMER
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- pheatmap vs heatmap.2
- PiCRUST2 Pipeline for Functional Prediction and Pathway Analysis in Metagenomics
- Should the inputs for GSVA be normalized or raw?
- Setup conda environments
- Kraken2 Installation and Usage Guide
- File format for single channel analysis of Agilent microarray data with Limma?
最新文章
- Somatic Variation Detection
- MicrobiotaProcess_PCA_Group3-4.R processing Data_Karoline_16S_2025
- Phyloseq.Rmd processing Data_Karoline_16S_2025
- Processing RNAseq_2025_WT_vs_ΔIJ_on_ATCC19606
最多评论文章
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- The top 10 genes
- Retrieving KEGG Genes Using Bioservices in Python
推荐相似文章
Enhanced Visualization of Gene Presence for the Selected Genes in Bongarts_S.epidermidis_HDRNA
QIIME + Phyloseq + MicrobiotaProcess (v1)